- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantitative Electron Microscopic Assay Using Random Sampling from Single Sections to Test Plastic Synaptic Changes in Hippocampus
(*contributed equally to this work) Published: Vol 8, Iss 15, Aug 5, 2018 DOI: 10.21769/BioProtoc.2946 Views: 6448
Reviewed by: Geoffrey C. Y. LauAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Single-Particle Tracking of AMPA Receptor-Containing Vesicles
Victor C. Wong [...] Erin K. O’Shea
Jun 5, 2025 2301 Views

Local Iontophoretic Application for Pharmacological Induction of Long-Term Synaptic Depression
Borys Olifirov [...] Pavel Belan
Jun 5, 2025 1939 Views

Ultrafast Isolation of Synaptic Terminals From Rat Brain for Cryo-Electron Tomography Analysis
Rong Sun and Qiangjun Zhou
Sep 5, 2025 3585 Views
Abstract
Studies over several decades on the organization of the CA1 hippocampus–a particularly favorable model for learning, memory and certain forms of cognition–have shown that the synaptic network in this brain region is plastic (Fortin et al., 2012). Recent evidence suggests that a number of environmental and endogenous stimuli may have a substantial effect on hippocampus-dependent cognitive function, implying enhanced synaptic plasticity in this brain region. Stimuli (e.g., food restriction, enriched environment, social interaction, gene-loss [knock-out animals], etc.) can trigger structural and functional plasticity (e.g., spine formation, increased expression of neurotrophic factors, synaptic function and neurogenesis) in the hippocampus (Stewart et al., 1989; Andrade et al., 2002; Babits et al., 2016). Using quantitative electron microscopy, we can study the synaptic neuropil of CA1 hippocampus in rodents during short- or long-term treatments and/or stimuli. Within the scope of this electron microscopic methodological construct, the density of various synaptic connections, the morphology and internal structure of excitatory spine synapses (e.g., the mean length and width of postsynaptic densities) can be quantified. Such quantitative ultrastructural measurement using high-resolution electron-microscopy may be applied to observe structural manifestations of synaptic plasticity in rodent brain tissue. The presented ultrastructural protocol may empower researchers to reveal details and synaptic changes which may not be obvious using only light microscopy. Ultrastructural data may provide substantial advances in our understanding of the changes in hippocampal synaptic architecture under different conditions.
Keywords: BrainBackground
We performed electron microscopy using random sampling from single sections to optimize sample size (over ~100 images per sample) and to detect changes in synaptic features in control and treated animals. The errors introduced by this approach are relatively modest (e.g., we found a numerical density of perforated spines in control animals similar to that of earlier studies also using single-section analysis (Calverley and Jones, 1987 and 1990). Our aim is not to make stereologically-rigorous estimates of the absolute values; instead, we want to determine whether there are significant differences between normal and treated animals. Since we use the same sampling method for all experimental groups, we should be able to detect any substantial changes induced by an appropriate treatment protocol. However, such random sampling has a potential to underestimate differences; other methods should also be used to confirm the impact of a given treatment on the hippocampus.
Materials and Reagents
- Pipette tips
- Aluminum foil
- Glass slides
- ACLAR® Fluoropolymer Films (Electron Microscopy Sciences, catalog number: 50425-10 )
- Glass engraving pen
- Sterile Scalpel Blades #11 (Electron Microscopy Sciences, catalog number: 72044-11 )
- Copper Grids (e.g., 300 mesh) (Sigma-Aldrich, catalog number: G4901 )
- Grid Storage Box, 100 Capacity (Electron Microscopy Sciences, catalog number: 71140 )
- Parafilm two-way stretch 100 mm x 38 m long (TAAB, catalog number: P069 )
- Glass vials with PE stoppers (Fisher Scientific, catalog number: 03-339-26C)
Manufacturer: DWK Life Sciences, catalog number: KFS60965D2 . - Glass beakers
- Whatman® qualitative filter paper, Grade 1 (Sigma-Aldrich, catalog number: WHA1001150 )
- Cyanoacrylate superglue
- Paraformaldehyde (4% in PB) (powder) (Sigma-Aldrich, catalog number: 158127 )
- Glutaraldehyde (Electron Microscopy Sciences, catalog number: 16310 )
- NaH2PO4·H2O (Sigma-Aldrich, catalog number: S9638 )
- Na2HPO4·7H2O (Sigma-Aldrich, catalog number: S2429 )
- HCl (Sigma-Aldrich, catalog number: 30721 )
- NaOH (Sigma-Aldrich, catalog number: S2770 )
- Osmium tetroxide solution (2% in H2O) (Sigma-Aldrich, catalog number: 75633 )
Notes:- This stock solution can be diluted with 0.2 M PB to reach the final 1% working concentration used for contrasting.
- Extremely toxic, handle only under an approved fume hood, wear proper protective clothing, handle with care; do not dispose of in sinks; follow local regulation for toxic waste disposal.
- This stock solution can be diluted with 0.2 M PB to reach the final 1% working concentration used for contrasting.
- Ethanol, absolute (diluted with distilled water as requested by Procedure) (Merck, catalog number: 1009831011 )
- Uranyl acetate (Electron Microscopy Sciences, catalog number: 22400 )
- Propylene oxide (Sigma-Aldrich, catalog number: 82320 )
Note: Extremely toxic, handle only under an approved fume hood, wear proper protective clothing, handle with care; do not dispose of in sinks; follow local regulations for toxic waste disposal. - DurcupanTM Resin set (Sigma-Aldrich, catalog number: 44610 )
- Lead citrate solution (Leica Ultrostain II, TAAB, Leica Microsystems, catalog number: T534/2 )
Notes:- The 3% lead citrate solution is pre-packed in vacuum tight bags under helium atmosphere.
- Toxic, handle with care and collect after use; follow local regulation for toxic waste disposal.
- The 3% lead citrate solution is pre-packed in vacuum tight bags under helium atmosphere.
- 0.1 M Phosphate buffer (PB, pH 7.4) (see Recipes)
- 4% of formaldehyde solution with 0.2% of glutaraldehyde (see Recipes)
- DurcupanTM ACM resin (See Recipes)
Equipment
- Stereo (dissecting) microscope (Leica Microsystems, model: Leica S6D )
- Vibratome (e.g., Leica Biosystems, model: Leica VT 1000S )
- LSETM Incubator digital shaker (Corning, catalog number: 6781-4 )
- Refrigerator
- Fume hood
- Thermostat
- Ultramicrotome (Leica Microsystems, Reichert, model: Reichert Ultracut S )
- Diamond knife (Diatome ultra 45°, at least 1.5 mm wide)
- Perfect loop for section pick-up (Electron Microscopy Sciences, catalog number: 70944 )
- Transmission electron microscope, operating at 80 kV (e.g., JEOL, model: JEM-1011 )
- Circular-top Analog Hotplate Stirrer (DLAB MS-H-S, DLAB Instruments Ltd.)
- Forceps suitable for grid-handling
- Freezer
Software
- NIH ImageJ (https://imagej.nih.gov/ij/)
- Kaleidagraph (Synergy Software http://www.synergy.com/wordpress_650164087/kaleidagraph/)
- AnalySIS from Soft Imaging System (Münster, Germany)
Procedure
- Using a brain stereotaxic atlas as a guide (e.g., Allen Brain Atlas http://mouse.brain-map.org/static/atlas) under a stereomicroscope cut general area from the intact brain, fixed with at least 2% PFA and variable proportion of glutaraldehyde (at least 0.1% up to as much as 2%). Continue to cut rodent brain under the stereomicroscope into a hemisphere wedge including the hippocampus.
- Mount the brain sample on vibratome plate with any type of cyanoacrylate superglue (Figure 1). Use a small drop (~50 μl) of superglue only, and spread it with a plastic pipette tip, so it covers the same area as the size of the brain block. Simply put the brain chunk onto the glue-covered area (do not apply pressure, nor let the brain dry out!). Once the tissue block is on the glued plate, move it quickly into the 0.1 M PB solution-filled chamber of the vibratome. The glue will rapidly polymerize, and sectioning can be started immediately.

Figure 1. Coronal brain hemisphere wedge mounted on vibratome plate with superglue - Cut 50-90 µm thin free-floating sections with a vibratome (see Video 1). The chamber of the vibratome should be filled with 0.1 M PB, so the brain sample submerges beneath the surface of the liquid. The settings of the vibratome should be optimized to the actual machine in use, and are highly dependent on the condition of the tissue. For robustly fixed material, higher speed and lower frequency should be used. For weakly fixed material, lower cutting speed and higher frequency may be optimal. Video 1. Vibrating blade microtome sectioning brain block. Semi-automatic sectioning at high-speed; after programmed sectioning window which ensures extremely quick sectioning of the brain tissue block sample.
- Collect free-floating sections in glass vials (using a round bristle paint brush) and incubate them on a mildly moving shaker in the following solutions:
- Incubate for 2 x 5 min in 0.1 M PB, pH 7.4.
- Incubate for 1 h in 1% Osmium tetroxide in 0.1 M PB, pH 7.4 (toxic, wear protective clothing, under a hood).
- Incubate for 2 x 5 min in 0.1 M PB, pH 7.4 (Stopping point, the sample can be stored in a refrigerator overnight at this step).
- Incubate for 5 min in 30% ethanol.
- Incubate for 5 min in 50% ethanol.
- Incubate for 30 min in 1% Uranyl acetate in 70% ethanol.
Notes:- This incubation step (Step 4f) is light-sensitive; the solution should be protected from light by wrapping with aluminum foil.
- Uranyl acetate is toxic. Wear protective clothing, work under fume hood, and dispose of according to local regulations.
- This incubation step (Step 4f) is light-sensitive; the solution should be protected from light by wrapping with aluminum foil.
- Incubate for 5 min in 80% ethanol.
- Incubate for 5 min in 95% ethanol.
- Incubate for 2 x 5 min in 100% ethanol.
- Incubate for 5 min in 1:1 100% ethanol/Propylene oxide.
- Incubate for 2 x 5 min in 100% Propylene oxide.
- Incubate for 30 min in 1:1 Propylene oxide/Resin
- Incubate for 30 min in 1:3 Propylene oxide/Resin.
- Incubate for 2 x 1 h in resin (Stopping point, the sample can be stored in a refrigerator overnight in either resin step).
- Incubate for 2 x 5 min in 0.1 M PB, pH 7.4.
- Waferize: create a multi-layered sandwich composed of 5 layers:
1) Glass slide;
2) A rectangular ACLAR® film, that is 1-2 mm bigger in each direction, than the glass slide (this can be cut manually with scissors from the Letter or A4-sized film);
3) The osmicated, resin infiltrated sections can be directly transferred onto the ACLAR® film with a bit of extra resin. Once all the sections are arranged on the film;
4) Cover them with another ACLAR® film, same size as the one beneath the tissue samples, avoid the formation of bubbles;
5) Scratch the name/ID of your samples onto the glass slide that covers the entire ‘wafer’ with a diamond point glass engraving pen and polymerize in an appropriate thermostat at 60 °C for 36-48 h. - After cured, remove upper glass slide and upper ACLAR® film, cut out the area of interest with No. 11 scalpel blade (Figure 2).
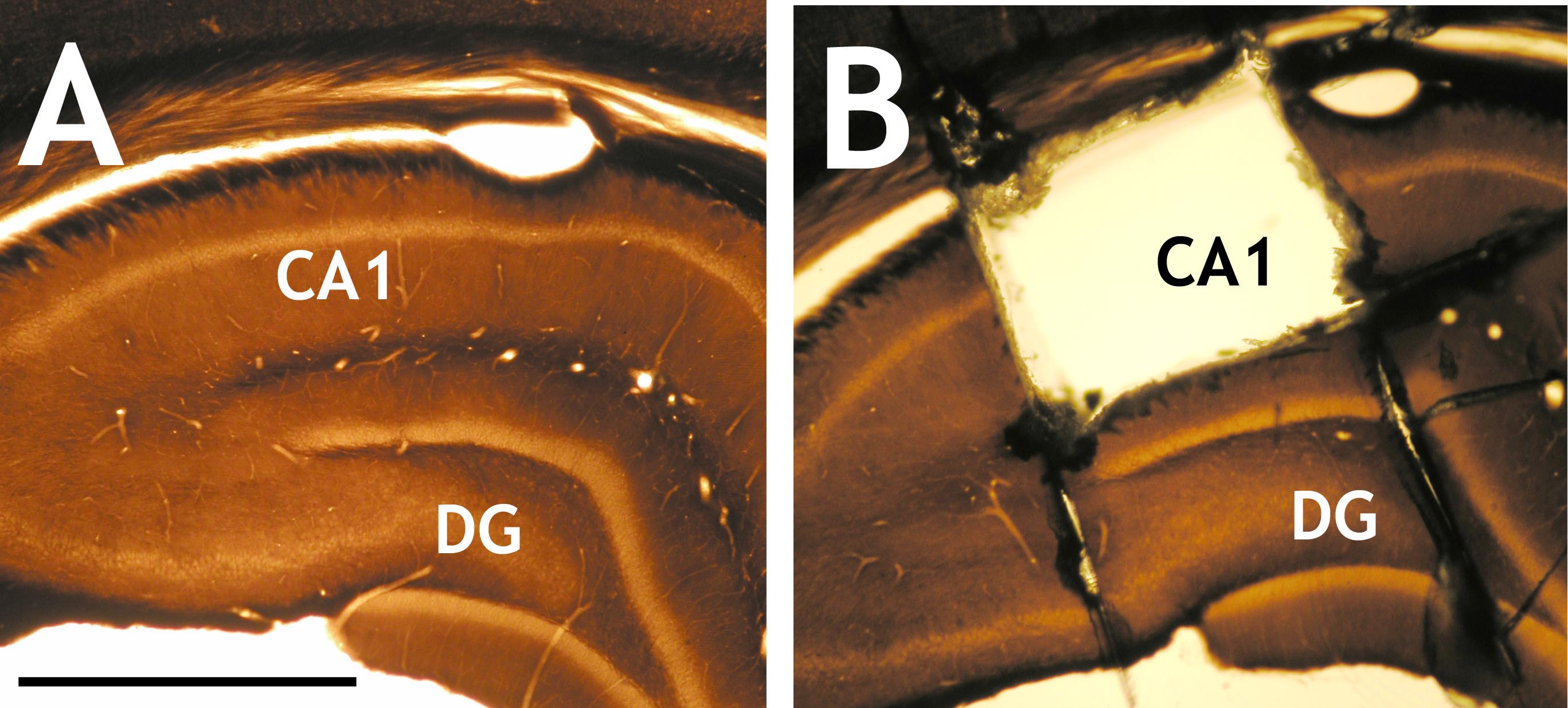
Figure 2. Osmium-fixed and resin embedded brain sections. A. Image showing osmium-fixed and resin embedded brain section with the hippocampal proper; B. Image showing the sampling area of CA1 (Cornu Ammonis 1) region, which was cut out with a scalpel blade under a dissecting microscope. DG: Dentate gyrus. Scale bar = 1 mm. - Mount on a resin block with super glue.
- Cut 50-70 nm thin ultrathin sections with an ultramicrotome (Figure 3).
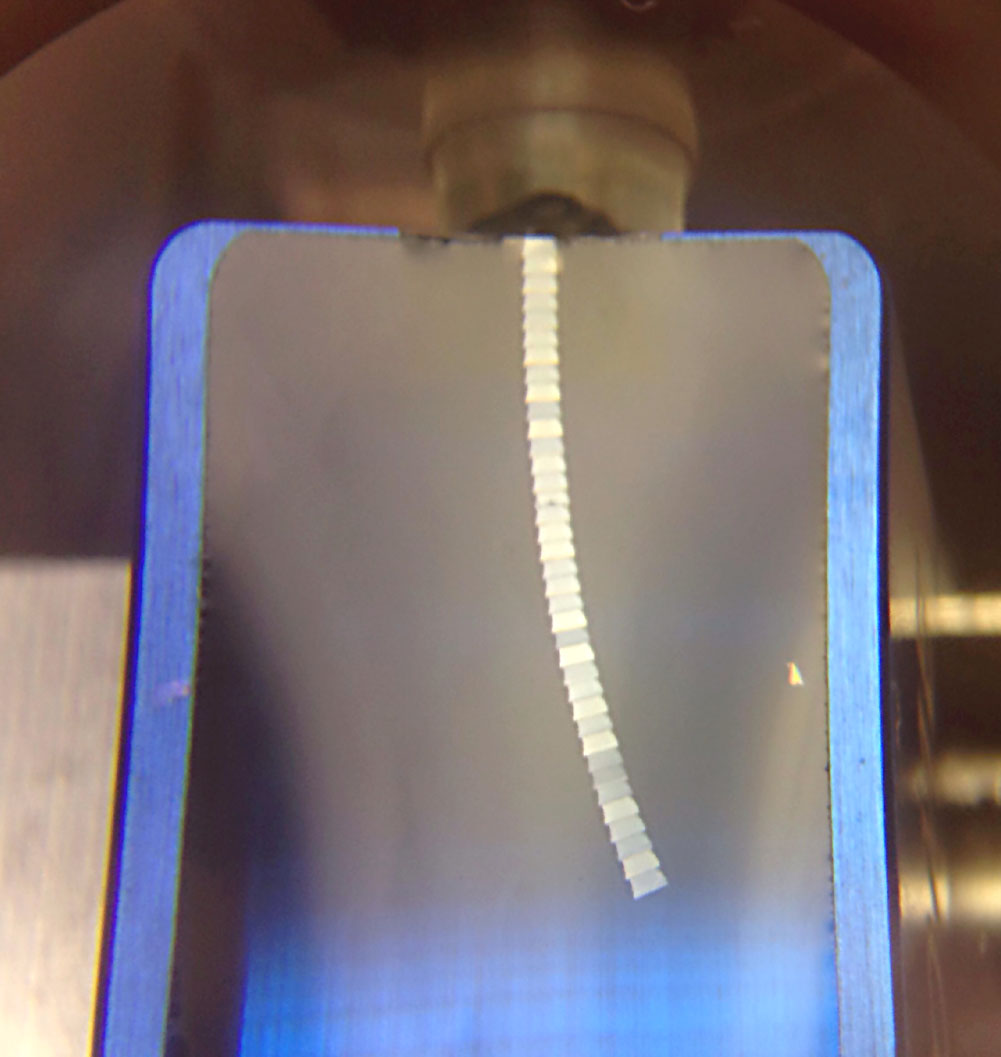
Figure 3. Image showing floating 50-70 nm ultrathin CA1 sections forming a ribbon in the distilled-water filled tank of a DIATOME ultra 45° 1.5 mm diamond knife (blue) - Mount sections on e.g., 300 mesh copper grids with the help of a perfect loop. See details at the vendor’s website (Perfect Loop for Ultra Thin sections).
- Collect grids into a storage box.
- Use Leica’s Ultrostain II lead citrate solution to make ~50 μl droplets in a Petri dish covered by Parafilm, and incubate the grids within these droplets for 1 min.
- Eliminate traces of the contrasting lead solution by quickly (and gently) moving the grids in three consecutive dH2O filled glass vial.
- Examine sections with a transmission electron microscope.
Data analysis
Data collection and analysis
- A digital camera (e.g., Mega-View-III digital camera and a Soft Imaging System (SIS, Münster, Germany) should be used for the computerized acquisition of the electron micrographs. Electron micrographs should be obtained according to the settings of the given TEM; in general, 80 KV should be used for the optimal contrast of brain tissue. Five to ten sections should be analyzed per block, and two blocks per animal may be used to collect micrographs. Sample areas (at least 50 µm2 per animal) may be chosen in a pseudo-random fashion and photographed at a uniform magnification (e.g., final magnification, 20,000x, with which our camera and scope settings provide ~33 μm2 sized fields).
- Postsynaptic dendritic spines, axonal boutons, multi-synaptic boutons (MSB; a single presynaptic bouton that forms separate synapses with multiple spine heads), spines with perforated postsynaptic density (PSD) and profiles of mitochondria can be identified on electron micrographs (Figure 4). Various profiles, e.g., spine and mitochondrion profile area, circularity (4π*area/perimeter2; a value of 1.0 indicates a perfect circle, and 0 indicates a completely flattened shape), and the length of the PSD can be measured using the engine provided by NIH: ImageJ v1. (Schneider et al., 2012). Profiles of spines, terminals, mitochondria must be identified and their parameters should be manually measured (Figure 5) with the built in engine of ImageJ (use the tool ‘Shape descriptors’ to record these values).

Figure 4. Representative electron micrographs of synaptic neuropil in the middle ⅓ of stratum radiatum of CA1 hippocampus, showing pseudocolored postsynaptic spines (orange) and in material from mouse brain with different treatment conditions (1 and 2). Arrowheads indicate perforated spine-synapses, which are more frequent in the hippocampus of “Condition 2”. Scale bar = 1 μm.
Figure 5. Manual measurements of profiles in an electron micrograph using ImageJ. Panel indicates the way a single spine profile may be selected and measured (thin arrow, yellow outline), and how the data is generated by the engine of Image J (thick arrow). - Data may be compiled using Kaleidagraph (Synergy Software, Reading, PA, USA) or any other similar software. The means and the possible effects of treatment can be determined between groups by one-way ANOVA with Bonferroni and Tukey’s post-hoc tests to analyze differences between treatments, with a P < 0.05 considered statistically significant. To eliminate bias, data collection and quantification should always be performed blindly.
Recipes
- Phosphate Buffer for 1 L of solution (0.1 M pH 7.4)
- Add 800 ml of distilled water in a glass beaker
- Add 20.209 g of Sodium phosphate dibasic to the solution
- Add 3.394 g of Sodium phosphate monobasic to the solution
- Fill up to 1 L
- Check and adjust the solution to final desired pH using HCl or NaOH
- Add 800 ml of distilled water in a glass beaker
- 4% of formaldehyde solution with 0.2% of glutaraldehyde (1 L)
- Add 600 ml of 0.1 M PB into a glass beaker on a heated stir plate in a ventilated hood
- Heat while stirring to approximately 60 °C. Please take care not to overheat the solution
- Add 40 g of paraformaldehyde powder to the heated PB solution. The powder will dissolve slowly into solution
- While still stirring, slowly add 1 M NaOH solution dropwise with a pipette until the solution is clear
- Once the paraformaldehyde is dissolved, cool the solution and filter through filter paper to remove coarse particles and precipitates
- Finally, adjust the volume of the solution to 1 L with 0.1 M PB
- Store at 4 °C and add the glutaraldehyde immediately before use
- Add 600 ml of 0.1 M PB into a glass beaker on a heated stir plate in a ventilated hood
- DurcupanTM ACM resin
Notes:- DurcupanTM ACM resin is used for flat embedding of osmicated and dehydrated vibratome sections.
- Regarding the four DurcupanTM components (A, B, C and D), component C should be kept refrigerated, the rest at room temperature.
- Mix 10 g of A and 10 g of B components. Add 0.3 g of C and 0.3 g of D components and stir to help dissolve with a magnetic stirrer. Stirring must be thorough and slow to avoid bubble build up. Cover the mixture while stirring to avoid excess exposure to moisture
- After 30 min of stirring at room temperature, let the mixture rest for an hour. After this time it is ready to use. Excess resin can be kept in the freezer (-18 °C) for a week
- DurcupanTM ACM resin is used for flat embedding of osmicated and dehydrated vibratome sections.
Acknowledgments
Authors declare no conflict of interest. We thank Tünde Magyar and Renáta Pop for excellent technical assistance, help with animal handling, experiments and data collection. The Project was supported by the European Union and co-financed by the European Social Fund (grant agreement No. EFOP-3.6.2- 16-2017- 00008; title: The role of neuro-inflammation in neurodegeneration: from molecules to clinics. B.R. is also supported by the János Bolyai Research Fellowship of the Hungarian Academy of Sciences. This protocol has been adapted and modified from the following works: Hazai et al., 2013; Kim et al., 2015, and Babits et al., 2016.
References
- Andrade, J. P., Lukoyanov, N. V. and Paula-Barbosa, M. M. (2002). Chronic food restriction is associated with subtle dendritic alterations in granule cells of the rat hippocampal formation. Hippocampus 12(2): 149-164.
- Babits, R., Szoke, B., Sotonyi, P. and Racz, B. (2016). Food restriction modifies ultrastructure of hippocampal synapses. Hippocampus 26(4): 437-444.
- Calverley, R. K. and Jones, D. G. (1987). Determination of the numerical density of perforated synapses in rat neocortex. Cell Tissue Res 248(2): 399-407.
- Calverley, R. K. and Jones, D. G. (1990). Contributions of dendritic spines and perforated synapses to synaptic plasticity. Brain Res Brain Res Rev 15(3): 215-249.
- Fortin, D. A., Srivastava, T. and Soderling, T. R. (2012). Structural modulation of dendritic spines during synaptic plasticity. Neuroscientist 18(4): 326-341.
- Hazai, D., Szudoczki, R., Ding, J., Soderling, S. H., Weinberg, R. J., Sótonyi, P. and Rácz, B. (2013). Ultrastructural abnormalities in CA1 hippocampus caused by deletion of the actin regulator WAVE-1. PLoS One 8(9): e75248.
- Kim, I. H., Rossi, M. A., Aryal, D. K., Racz, B., Kim, N., Uezu, A., Wang, F., Wetsel, W. C., Weinberg, R. J., Yin, H. and Soderling, S. H. (2015) Spine pruning drives antipsychotic-sensitive locomotion via circuit control of striatal dopamine. Nat Neurosci 18(6): 883-891.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Stewart, J., Mitchell, J. and Kalant, N. (1989). The effects of life-long food restriction on spatial memory in young and aged Fischer 344 rats measured in the eight-arm radial and the Morris water mazes. Neurobiol Aging 10(6): 669-675.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Marcello, G. M., Szabó, L. E., Sotonyi, P. T. and Rácz, B. (2018). Quantitative Electron Microscopic Assay Using Random Sampling from Single Sections to Test Plastic Synaptic Changes in Hippocampus. Bio-protocol 8(15): e2946. DOI: 10.21769/BioProtoc.2946.
Category
Neuroscience > Neuroanatomy and circuitry > Cortex
Neuroscience > Cellular mechanisms > Synaptic physiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link