- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measuring CD38 Hydrolase and Cyclase Activities: 1,N6-Ethenonicotinamide Adenine Dinucleotide (ε-NAD) and Nicotinamide Guanine Dinucleotide (NGD) Fluorescence-based Methods
(*contributed equally to this work) Published: Vol 8, Iss 14, Jul 20, 2018 DOI: 10.21769/BioProtoc.2938 Views: 9376
Reviewed by: Andrea PuharYONG TENGAnca Savulescu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of the Activity of Wildtype and Disease-Causing ALPK1 Mutants in Transfected Cells With a 96-Well Format NF-κB/AP-1 Reporter Assay
Tom Snelling
Nov 20, 2024 1633 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2323 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 531 Views
Abstract
CD38 is a multifunctional enzyme involved in calcium signaling and Nicotinamide Adenine Dinucleotide (NAD+) metabolism. Through its major activity, the hydrolysis of NAD+, CD38 helps maintain the appropriate levels of this molecule for all NAD+-dependent metabolic processes to occur. Due to current advances and studies relating NAD+ decline and the development of multiple age-related conditions and diseases, CD38 gained importance in both basic science and clinical settings. The discovery and development of strategies to modulate its function and, possibly, treat diseases and improve health span put CD38 under the spotlights. Therefore, a consistent and reliable method to measure its activity and explore its use in medicine is required. We describe here the methods how our group measures both the hydrolase and cyclase activity of CD38, utilizing a fluorescence-based enzymatic assay performed in a plate reader using 1,N6-Ethenonicotinamide Adenine Dinucleotide (ε-NAD) and Nicotinamide Guanine Dinucleotide (NGD) as substrates, respectively.
Keywords: CD38Background
Current studies on age-related development of metabolic dysfunction and frailty are each day in more evidence. It is known that, as the aging progresses, the NAD+ levels decrease in an expected process. Recent studies have shown that a reduction in nicotinamide adenine dinucleotide (NAD+) is implicated in the development of age-associated metabolic decline (Massudi et al., 2012). Increased NAD+ levels in vivo, results in activation of pro-longevity and health span-related factors and improves several physiological and metabolic parameters of aging (Camacho-Pereira et al., 2016), including muscle function, exercise capacity, glucose tolerance, and cardiac function in mouse models of natural and accelerated aging.
Due to its role in NAD+ metabolism, the study of CD38 and its functions has been of great importance. CD38 was first identified in 1980 as a structural cell surface marker for the characterization of immune cells (Malavasi et al., 2008; van de Donk et al., 2016), and its first association as a NAD hydrolase enzyme was in the following decade (Kontani et al., 1993). However, during the past years, its enzymatic activities were more clearly elucidated. Initially, CD38 has been implicated to be responsible for the synthesis of the second messengers, cyclic ADP-ribose (cADPR), ADPR and nicotinic acid–adenine dinucleotide phosphate (NAADP) (Chini et al., 2002). These products are involved in calcium signaling and control many biological processes including lymphocyte proliferation and insulin secretion (Kato et al., 1999). However, its major enzymatic activity is the NAD+ hydrolysis, placing CD38 as the major NADase in several mammalian tissues and as an important regulator of NAD+-dependent processes (Aksoy et al., 2006).
The primary catalytic reaction of CD38 involves the cleavage of the high energy β-glycosidic bond between nicotinamide and ribose. During catalysis, the removal of the nicotinamide from β-NAD is coupled with the formation of intermediates that are stabilized through H-bonds between their ribosyl groups and the catalytic residue Glu226, a residue required for the NADase and cyclase activity of the enzyme (Sauve et al., 2000; Liu et al., 2009). These intermediates are released from the catalytic site forming ADPR or cADPR (Figure 1). In general, the majority of the CD38 NADase catalytic activity will generate nicotinamide, but also ADPR and cADPR which have been shown to have second messenger signaling roles through the activation of ryanodine receptor (RYR2). The full description of these pathways and Ca2+ signaling can be found in the remarkable works of Galione (1994) and Chini and Dousa (1996). The roles of CD38 as a cyclase and of NAD-derived calcium messengers in physiology and pathology have been extensively reviewed (Sauve et al., 2000; Chini, 2009).
Physiologically, CD38 has been implicated in the regulation of metabolism and the pathogenesis of the aging process, and of multiple conditions, such as obesity, diabetes, heart disease, asthma and inflammation. Therefore, the study of CD38, its activities, and possible modulators are of great interest. Our protocol presents a method of measuring its hydrolase and cyclase activities, utilizing ε-NAD and NGD techniques (Graeff et al., 1994) with a fluorescence-based enzymatic assay performed in a plate reader, in a consistent and reproducible manner (Figure 2).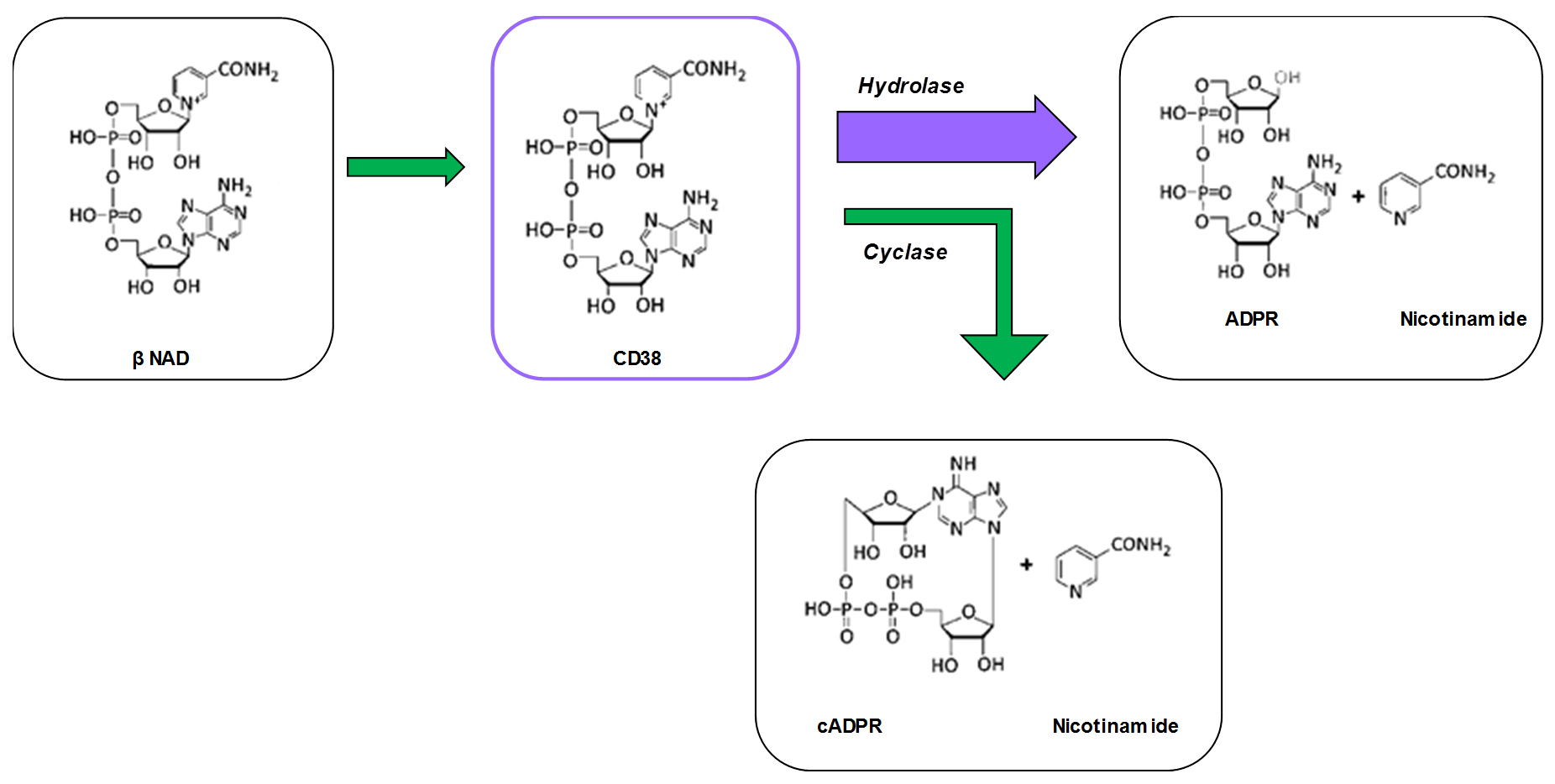
Figure 1. Schematic illustrating the reactions catalyzed by CD38 under physiological conditions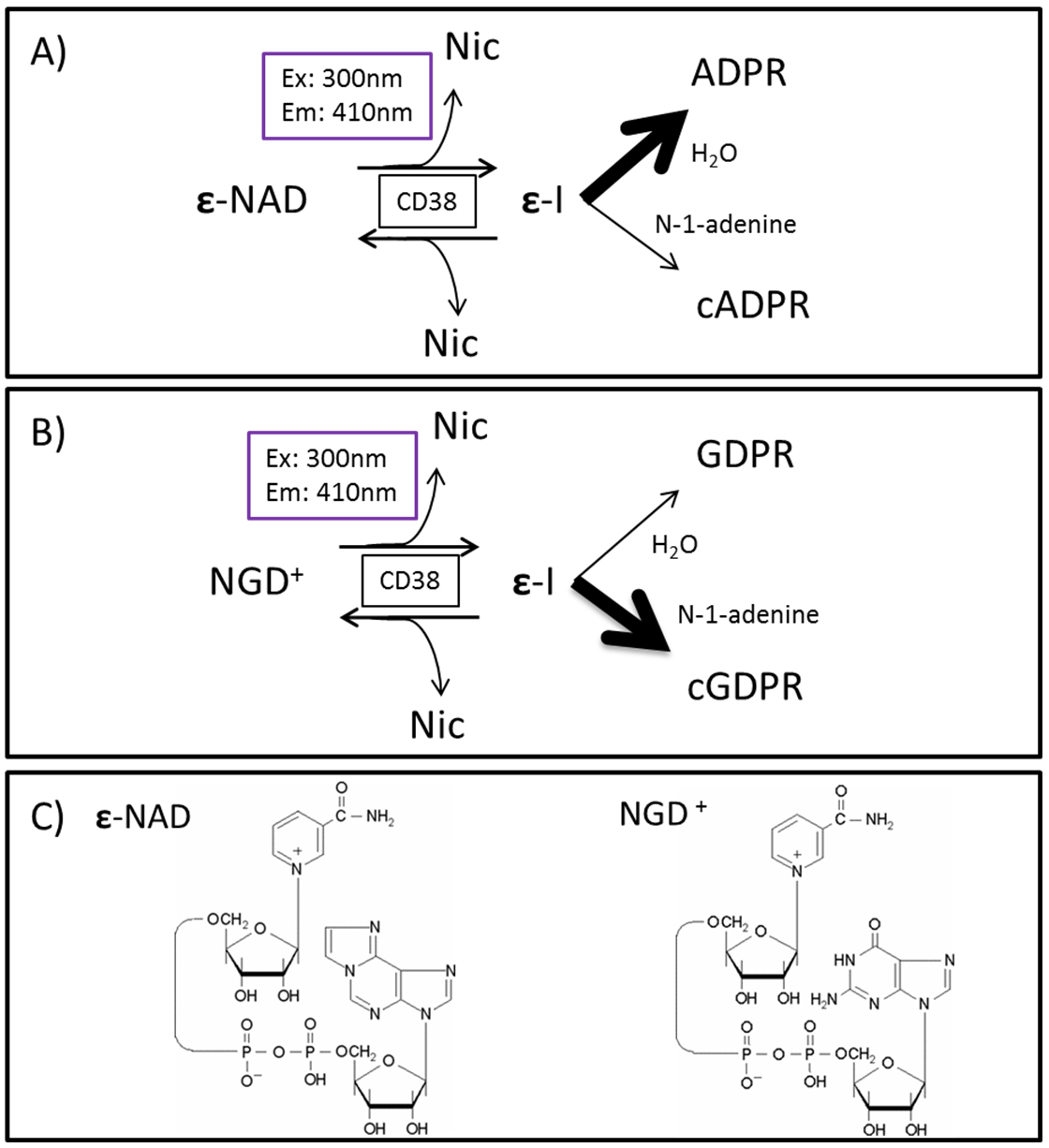
Figure 2. CD38 and substrate schematics. A. CD38 hydrolase activity (ε-NAD as substrate); B. Cyclase activity (NGD as substrate); Strong arrow indicates which product is preferentially formed in each reaction. C. Molecular structure of ε-NAD and NGD. *Nic = Nicotinamide, ε-I = enzyme-intermediate complex, (c)ADPR = (cyclic) ADP-ribose, (c)GDPR = (cyclic) GDP-ribose, Ex = excitation wavelength, Em = emission wavelength.
Materials and Reagents
- Plastic tips 1,000 (Thermo Scientific®, Molecular Bioproducts, catalog number: 3101 )
- Plastic tips 200 (Thermo Fisher Scientific, Molecular Bioproducts, catalog number: 3551 )
- 96-well plate (Microfluor 1 White flat-bottom plate) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 7705 )
- 1.5 ml tubes
- 60 mm culture dishes (Fisher Scientific, FisherbrandTM, catalog number: FB012921 )
- Tissues of interest: any tissue can be used to measure NAD+/NADH levels
- Cells of interest: we usually use A549, JURKAT, Patu 9888T to measure NAD+/NADH levels
- Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A7906 )
- Sucrose (Sigma-Aldrich, catalog number: S0389-1KG )
- Tris Base (Trizma® base, Sigma-Aldrich, catalog number: T6066 )
- Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, catalog number: 5000006 )
- CD38 human recombinant enzyme (R&D Systems, catalog number: 2404-AC-010 )
- CD38 inhibitor (Merck, Calbiochem, catalog number: 538763 )
- Anti-CD38 antibody–Isatuximab (Creative-Biolabs, catalog number: TAB-432CQ )
- Nicotinamide guanine dinucleotide sodium salt (NGD) (Sigma-Aldrich, catalog number: N5131 )
- Nicotinamide 1, N6-ethenoadenine dinucleotide (ε-NAD) (Santa Cruz Biotechnology, catalog number: sc-215559 )
- MES (Sigma-Aldrich, catalog number: M3671 )
- Sodium chloride (Sigma-Aldrich, catalog number: S7653 )
- Nanopure water
- HCl
- Sucrose Buffer (see Recipes)
- rhCD38 enzyme buffer (see Recipes)
Materials necessary to collect cells:
- Scraper–Corning cell lifter (Corning, catalog number: 3008 )
- Trypsin-EDTA 0.25% (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056 )
- Phosphate Buffered Saline (PBS) 1x (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
Equipment
Note: The brands and models indicated are the ones used by our group, similar equipment can be used as well.
- Pipettes (10, 200, 1,000 μl)
- Scissors
- Graduated cylinder
- Repeat pipette (Eppendorf, model: Repeater® M4 )
- Scale (Mettler-Toledo International, model: AG104 )
- Microcentrifuge (Eppendorf, model: 5424 )
- Homogenizer (Tissue Tearor, Bio Spec Products, catalog number: 780CL-04 )
- Sonic Dismembrator (Fisher Scientific, model: Model 100 Sonic Dismembrator)
- Spectrophotometer (BioTek Instruments, model: Epoch 2 )
- Vortex (Scientific Industries, model: Vortex-Genie 2 , catalog number: G560)
- Plate reader (Molecular Devices, model: SpectraMax Gemini XPS )
Software
- Gen5 Microplate Reader and Imager Software (BioTek Instruments)
- Microsoft Excel (Microsoft Corporation)
- SoftMax Pro 6 (Molecular Devices, LLC)
- GraphPad Prism 7 (GraphPad Software, Inc)
Procedure
The CD38 activity assay can be done mainly with three different sources of CD38, cells, tissues, and recombinant enzyme. We are going to describe the procedure for cells and tissues first, and then describe the procedure using recombinant CD38 enzyme. The assay is performed in a 96-well white opaque plate (refer to Notes for plate information), at least in duplicates, with a final volume of 200 μl per well.
- Cell and tissue samples
It is necessary to lyse the cells and tissue samples to be able to measure CD38 activity. To do so, we lyse the samples in sucrose buffer with sonication. The optimal volume of buffer should be empirically determined for each cell type and pellet size, as well as tissue weight, to ensure efficient lysis and an optimal final concentration of proteins in the lysate. We normally use 300 μl for 20 mg of tissue, and 100 μl of sucrose buffer if using cells from a 60 mm dish (2-3 x 106 cells). Keep buffer and samples on ice during all the assay steps.- Sample preparation
- Cells: wash cells in PBS, collect them according to standard procedures suitable for each cell type (adherent cells via scraping or trypsinization; cells that grow in suspension via transferring and pelleting) in 1.5 ml tubes and pellet them by centrifugation (30 sec, at 11.7 x g). Aspirate supernatant, re-suspend cells with 100 μl of sucrose buffer, if using 2-3 x 106 cells) and sonicate the samples at 30-50 W of power and 20 kHz of frequency on ice, for 3 times of 5 sec each.
- Tissue: in a 2 ml tube, add the piece of tissue (approx. 20 mg), add 300 μl of sucrose buffer and homogenize with scissors and a mechanical homogenizer until there are no visible chunks. Then, sonicate as explained above for cells. Remember to keep samples on ice during this process to avoid that the heat generated degrades the CD38 enzyme.
After sonicating, centrifuge samples for 10 min at 13.8 x g, and at 4 °C. Transfer supernatant to a new tube for protein measurement by the Bio-Rad protein assay, and following steps. Discard pellet.
- Cells: wash cells in PBS, collect them according to standard procedures suitable for each cell type (adherent cells via scraping or trypsinization; cells that grow in suspension via transferring and pelleting) in 1.5 ml tubes and pellet them by centrifugation (30 sec, at 11.7 x g). Aspirate supernatant, re-suspend cells with 100 μl of sucrose buffer, if using 2-3 x 106 cells) and sonicate the samples at 30-50 W of power and 20 kHz of frequency on ice, for 3 times of 5 sec each.
- Normalizing samples
Normalize samples with sucrose buffer to obtain a mass of 20-100 μg of protein if tissue is the source of enzyme, or 50-100 μg of protein per well if cells are being used. The volume of sample to be pipetted into each well is 100 μl. For tissues with high CD38 expression, like spleen, aim to the lower end of the interval. Proceed to “Prepare reaction mix” step.
- Sample preparation
- Recombinant CD38 enzyme
If using commercially available recombinant CD38 enzyme, the preparation of the assay differs from how it is done with cells and tissues. The total volume in the well is maintained at 200 μl, the reaction mix is the same (100 μl) but the volume of enzyme mix can vary if one wants to test an inhibitor or activator. This method can also be adapted to use with cells or tissue samples, although our group normally treat cells or animals previously with inhibitors/activators and proceed as described above.- Prepare test compounds/inhibitors
Prepare a dilution of the test compounds 4x of the desired final concentration in sucrose buffer to a volume of 50 μl per well. Also, prepare a blank sample, with 50 μl of sucrose buffer and no test compounds. We recommend the use of a known CD38 inhibitor, such as 78c (50 nM final), or anti-CD38 antibodies as a control for the CD38 activity and test compounds. For human samples or human recombinant CD38 enzyme we suggest the use of Isatuximab (1 μg/ml), a well-known monoclonal anti-CD38 antibody already in Phase 3 clinical trials for Multiple Myeloma. - Prepare enzyme mix
The enzyme mix, which contains Recombinant enzyme (10 ng/μl), BSA (40 mg/ml in water) and sucrose buffer, will have a total volume of 50 μl per well. We suggest an incubation period of 15 min at room temperature if antibodies are being used, based on our experience. For 78c no incubation is required, since this molecule only binds to the enzyme-substrate product. Calculate the number of wells that are going to be used and use the following proportion to prepare the enzyme mix: Recombinant enzyme 1 μl/well; BSA 4 μl/well; and sucrose buffer 45 μl/well.
Note: The following steps are common for both samples type.- Prepare reaction mix
- Based on which CD38 activity one wants to measure, two different substrates are added to sucrose buffer. For the hydrolase/NADase activity, ε-NAD is used. For the cyclase activity, NGD is the substrate of choice.
- Prepare the reaction mix based on the number of wells to be used, considering 100 μl of reaction mix per well. For each 1 milliliter of total reaction mix, 5 μl of 10 mM ε-NAD and 40 μl of 10 mM NGD should be added, in order to achieve a final concentration of 50 and 200 μM, respectively. Vortex the tube and leave it at room temperature.
- Based on which CD38 activity one wants to measure, two different substrates are added to sucrose buffer. For the hydrolase/NADase activity, ε-NAD is used. For the cyclase activity, NGD is the substrate of choice.
- Setting up the reaction
- Before start pipetting samples to the plate, make sure that the plate reader is properly configured. Set up machine to read fluorescence at 300 nm excitation and 410 nm emission. Configure settings of plate type (96-well opaque), analysis type (kinetics), time of analysis (at least 1 h, readings every 30 sec), define area to be read on plate, and set it up to shake once for 5 sec before start reading.
- Finally, pipet 100 μl of normalized samples, or 50 μl of recombinant enzyme mix plus 50 μl of inhibitor/activator tested, at least in duplicates. It is advised to pipet sucrose buffer in a set of wells as a blank. Then, quickly add with a repeater pipette 100 μl of reaction mix to all wells, load plate on reader tray, and read.
- Before start pipetting samples to the plate, make sure that the plate reader is properly configured. Set up machine to read fluorescence at 300 nm excitation and 410 nm emission. Configure settings of plate type (96-well opaque), analysis type (kinetics), time of analysis (at least 1 h, readings every 30 sec), define area to be read on plate, and set it up to shake once for 5 sec before start reading.
- Prepare test compounds/inhibitors
Data analysis
Plot the values obtained in an X-Y graph. On the X-axis plot the time, and on the Y-axis plot the fluorescence unit values obtained during the reading process (Figure 3). For statistical analysis, we use GraphPad Prism. The enzymatic activity curves obtained can be compared to evaluate the effects of the compound studied in the CD38 enzyme. For instance, in Figure 3, the black curve “Buffer” indicates the enzyme function without a test compound (where sucrose buffer was added to the enzyme). The grey curve shows the effect of the CD38 inhibitor, 78c, clearly demonstrating that there was little increase in activity over time. The red and pink “Antibodies” curve shows the CD38 activity when an Anti-CD38 antibody, in two different concentrations, was added to the enzyme. The green curves represent two concentrations of an unknown test compound being evaluated for CD38 inhibition, in this case showing little effect on CD38 activity. “No enzyme” refers to wells where only sucrose buffer and reaction mix were added. Figures 3C and 3D show the raw data and graph of percentage of activity in a chosen time point for all the compounds previously described. To compare two data sets, we perform a Student’s t-test.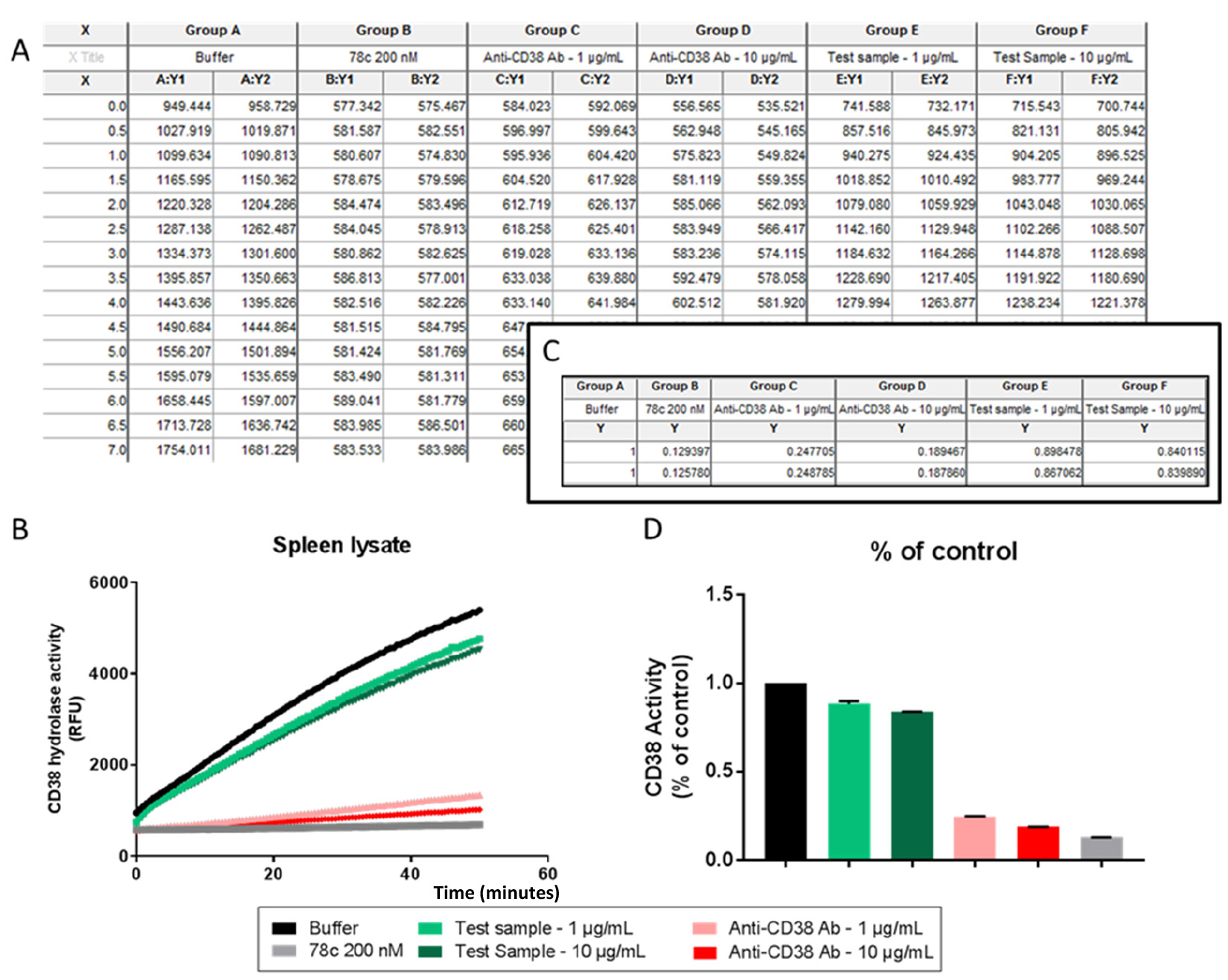
Figure 3. Data analysis on GraphPad Prism. A. Raw data; B. Final activity graph based on raw data analysis; C. Table of % of control values calculated per condition; D. Final % of control graph.
Notes
- It is important that in key experiments, results are also confirmed by following the degradation of NAD+, the natural substrate for both the CD38 NADase and cyclase enzymes, using HPLC to follow the hydrolysis of NAD+ to ADPR and cADPR (Aksoy et al., 2006).
- NADase activity can also be verified by HPLC analysis, performed by anion-exchange chromatography using an AG MP-1 column (Bio-Rad) eluted with a non-linear gradient of trifluoroacetic acid, as described previously (Aksoy et al., 2006). The nucleotides are detected by UV absorption at 254 nm, and the authenticity of NAD+ is confirmed by co-elution with NAD+ standards (Aksoy et al., 2006).
- We utilize 96-well standard opaque white plates for our assays, but we’ve obtained similar results with black plates, although they produce lower RFU values.
Recipes
- Sucrose Buffer (500 ml)
- Weigh 42.7875 g of sucrose and 2.4228 g of Tris base to achieve a concentration of 0.25 M Sucrose and 40 mM Tris
- Add 400 ml of Nanopure water, add the sucrose and Tris, adjust pH to 7.4 with HCl and complete the volume to 500 ml in a graduated cylinder
- Store at 4 °C up to 6 months
- Weigh 42.7875 g of sucrose and 2.4228 g of Tris base to achieve a concentration of 0.25 M Sucrose and 40 mM Tris
- rhCD38 enzyme buffer (5 ml)
- Weigh 24.4 mg of MES and 43.83 mg of NaCl to achieve a concentration of 25 mM MES and 150 mM NaCl
- Add 4 ml of Nanopure water, add the MES and NaCl, adjust pH to 6.5 with HCl and complete the volume to 5 ml in a graduated cylinder
- Weigh 24.4 mg of MES and 43.83 mg of NaCl to achieve a concentration of 25 mM MES and 150 mM NaCl
Acknowledgments
This work was supported in part by grants from the Ted Nash Long Life Foundation, the Glenn Foundation for Medical Research via the Paul F. Glenn Laboratories for the Biology of Aging at the Mayo Clinic, a grant from Calico Laboratories, the Mayo Foundation, National Institutes of Health (NIH) grants from the National Institute of Aging (NIA, grant AG-26094), the Pancreatic Cancer SPORE project from NIH/NCI to E.N.C (grant CA102701-08), the Mayo Clinic Center for Cell Signaling in Gastroenterology (NIDDK P30DK084567). Maria Auxiliadora-Martins was supported by a grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Brazil (#2016/20089-4).
Declaration of interests: Dr. Chini holds a patent on the use of CD38 inhibitors for metabolic diseases.
References
- Aksoy, P., White, T. A., Thompson, M. and Chini, E. N. (2006). Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun 345(4): 1386-1392.
- Camacho-Pereira, J., Tarrago, M. G., Chini, C. C. S., Nin, V., Escande, C., Warner, G. M., Puranik, A. S., Schoon, R. A., Reid, J. M., Galina, A. and Chini, E. N. (2016). CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab 23(6): 1127-1139.
- Chini, E. N. (2009). CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr Pharm Des 15(1): 57-63.
- Chini, E. N., Chini, C. C., Kato, I., Takasawa, S. and Okamoto, H. (2002). CD38 is the major enzyme responsible for synthesis of nicotinic acid-adenine dinucleotide phosphate in mammalian tissues. Biochem J 362(Pt 1): 125-130.
- Chini, E. N. and Dousa, T. P. (1996). Nicotinate-adenine dinucleotide phosphate-induced Ca2+-release does not behave as a Ca2+-induced Ca2+-release system. Biochem J 316 (Pt 3): 709-711.
- Galione, A. (1994). Cyclic ADP-ribose, the ADP-ribosyl cyclase pathway and calcium signalling. Mol Cell Endocrinol 98(2): 125-131.
- Graeff, R. M., Walseth, T. F., Fryxell, K., Branton, W. D. and Lee, H. C. (1994). Enzymatic synthesis and characterizations of cyclic GDP-ribose. A procedure for distinguishing enzymes with ADP-ribosyl cyclase activity. J Biol Chem 269(48): 30260-30267.
- Kato, I., Yamamoto, Y., Fujimura, M., Noguchi, N., Takasawa, S. and Okamoto, H. (1999). CD38 disruption impairs glucose-induced increases in cyclic ADP-ribose, [Ca2+]i, and insulin secretion. J Biol Chem 274(4): 1869-1872.
- Kontani, K., Nishina, H., Ohoka, Y., Takahashi, K. and Katada, T. (1993). NAD glycohydrolase specifically induced by retinoic acid in human leukemic HL-60 cells. Identification of the NAD glycohydrolase as leukocyte cell surface antigen CD38. J Biol Chem 268(23): 16895-16898.
- Liu, Q., Graeff, R., Kriksunov, I. A., Jiang, H., Zhang, B., Oppenheimer, N., Lin, H., Potter, B. V., Lee, H. C. and Hao, Q. (2009). Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase. J Biol Chem 284(40): 27637-27645.
- Malavasi, F., Deaglio, S., Funaro, A., Ferrero, E., Horenstein, A. L., Ortolan, E., Vaisitti, T. and Aydin, S. (2008). Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev 88(3): 841-886.
- Massudi, H., Grant, R., Braidy, N., Guest, J., Farnsworth, B. and Guillemin, G. J. (2012). Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One 7(7): e42357.
- Sauve, A. A., Deng, H., Angeletti, R. H. and Schramm, V. L. (2000). A covalent intermediate in CD38 is responsible for ADP-ribosylation and cyclization reactions. J Am Chem Soc 122(33): 7855-7859.
- van de Donk, N. W., Janmaat, M. L., Mutis, T., Lammerts van Bueren, J. J., Ahmadi, T., Sasser, A. K., Lokhorst, H. M. and Parren, P. W. (2016). Monoclonal antibodies targeting CD38 in hematological malignancies and beyond. Immunol Rev 270(1): 95-112.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
de Oliveira, G. C., Kanamori, K. S., Auxiliadora-Martins, M., Chini, C. C. S. and Chini, E. N. (2018). Measuring CD38 Hydrolase and Cyclase Activities: 1,N6-Ethenonicotinamide Adenine Dinucleotide (ε-NAD) and Nicotinamide Guanine Dinucleotide (NGD) Fluorescence-based Methods. Bio-protocol 8(14): e2938. DOI: 10.21769/BioProtoc.2938.
Category
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











