- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of TLR4 and CD14 Receptor Endocytosis Using Flow Cytometry
Published: Vol 8, Iss 14, Jul 20, 2018 DOI: 10.21769/BioProtoc.2926 Views: 10525
Reviewed by: Ivan ZanoniMeenal SinhaBenoit Stijlemans

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Macrophage Cellular Ferrous Iron (Fe2+) Content using a Highly Specific Fluorescent Probe in a Plate-Reader
Philipp Grubwieser [...] Christa Pfeifhofer-Obermair
Feb 5, 2024 2215 Views

Dissecting the Mechanical Control of Mitotic Entry Using a Cell Confinement Setup
Margarida Dantas [...] Jorge G. Ferreira
Mar 20, 2024 2732 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3547 Views
Abstract
After recognizing extracellular bacterial lipopolysaccharide (LPS), the toll-like receptor 4 (TLR4)-CD14 signaling complex initiates two distinct signaling pathways–one from the plasma membrane and the other from the signaling endosomes (Kagan et al., 2008). Understanding the early stages of TLR4 signal transduction therefore requires a robust and quantitative method to measure LPS-triggered TLR4 and CD14 receptor endocytosis, one of the earliest events of LPS detection. Here, we describe a flow cytometry-based method that we used recently to study the role of the ion channel TRPM7 in TLR4 endocytosis (Schappe et al., 2018). The assay relies on stimulating the cells with LPS and measuring the cell surface levels of TLR4 (or CD14) at various time points using flow cytometry. Although we detail the method specifically for TLR4 and CD14 from murine bone marrow-derived macrophages, it can be readily adapted to evaluate receptor endocytosis in a variety of other signaling contexts.
Keywords: Toll-like receptorBackground
Innate immune cells, including macrophages and dendritic cells, employ a variety of pattern recognition receptors (PRRs) to survey their environments for danger- and pathogen-associated molecular patterns. Trafficking and signaling of PRRs from various subcellular compartments enables wider immune surveillance and has emerged as an important design principle of innate immunity (Brubaker et al., 2015). The detection of the bacterial endotoxin LPS is highly dependent on TLR4 and its co-receptor CD14. The endocytosis of the TLR4 complex requires CD14 and is essential for LPS-induced macrophage activation (Zanoni et al., 2011; Tan et al., 2015). Endocytosis of TLR4 is essential to activate secondary signaling complexes at the newly-formed ‘signaling endosome,’ which promotes interferon regulatory factor 3-dependent transcription through the signaling adaptor TIR-domain containing adapter-inducing interferon-β (TRIF) (Kagan et al., 2008). TLR4 endocytosis has been observed in macrophages, dendritic cells, and epithelial cells (Roy et al., 2014). Understanding the underlying mechanisms of this critical step in macrophage activation requires a robust and quantitative method to measure LPS-triggered TLR4 endocytosis. Here, we describe a version of a flow cytometry-based method that was initially reported by Kagan and colleagues (Kagan et al., 2008), and used by others, to monitor TLR4 endocytosis. We have used the method recently to study the role of transient receptor potential melastatin-like 7 (TRPM7), an ion channel, in TLR4 endocytosis (Schappe et al., 2018). The experimental logic of this method relies on measuring the loss of TLR4 and CD14 staining at the cell surface after LPS treatment. We stain LPS-treated cells with an anti-TLR4 (or anti-CD14) fluorophore-conjugated antibody without permeabilization. The fluorescence intensity acquired using flow cytometry reports the relative quantity of receptor resident in the plasma membrane (Figure 1). Although specific for TLR4 and CD14, the assay can be readily adapted to evaluate receptor endocytosis in a variety of other signaling contexts.
Figure 1. Schematic of TLR4 and CD14 endocytosis protocol. Experimental workflow described in protocol “Procedure”.
Materials and Reagents
- Materials
- Pipette tips
- 5 ml round, disposable round-bottom tube (FACS Tube) (Corning, Falcon®, catalog number: 352052 )
- Aluminum foil (Genesee Scientific, catalog number: 88-101 )
- 0.2 μm bottle filter (Thermo Fisher Scientific, NalgeneTM, catalog number: 566-0020 )
- 6-well non-treated culture plates (Corning, catalog number: 3736 )
- Sterile cell scrapers (Fisher Scientific, FisherbrandTM, catalog number: 08-100-240 )
- Sterile individually packaged serological pipette (10 ml) (Greiner Bio One International, catalog number: 607160 )
- Sterile individually packaged serological pipette (5 ml) (Greiner Bio One International, catalog number: 606160 )
- 1.7 ml microfuge Eppendorf tubes (Genesee Scientific, Olympus Plastics, catalog number: 24-281 )
- NuncTM TripleFlaskTM Treated Cell Culture Flasks (Thermo Fisher Scientific, catalog number: 132867 )
- Falcon® 50 ml Conical Centrifuge Tube (Corning, catalog number: 352098 )
- Pipette tips
- Cell line
- L-929 cells (ATCC, catalog number: CCL-1 )
- L-929 cells (ATCC, catalog number: CCL-1 )
- Reagents
- LPS EB-Ultrapure (lipopolysaccharide from E. coli O111:B4, InvivoGen, catalog number: tlrl-3pelps )
- PBS (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- Mouse TruStain fcXTM (anti-CD16/32) (BioLegend, catalog number: 101320 )
- TLR4 [anti-mouse CD284] (PE) (clone: SA15-21; isotype: Rat IgG2a, κ) (BioLegend, catalog number: 145404 )
- CD14 [anti-mouse] (APC) (clone: Sa2-8; isotype: Rat IgG2a, κ) (Thermo Fisher Scientific, eBioscienceTM, catalog number: 17-0141-81 )
- RPMI 1640 (Thermo Fisher Scientific, GibcoTM, catalog number: 11875093 )
- Fetal bovine serum (heat-inactivated), certified, USA origin (Thermo Fisher Scientific, GibcoTM, catalog number: 10082147 )
- Trypan blue (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061 )
- HBSS, no calcium, no magnesium (Thermo Fisher Scientific, GibcoTM, catalog number: 14170112 )
- BSA (Bovine serum albumin) (Roche Molecular Systems, catalog number: 3116956001 )
- DMEM, high glucose (Thermo Fisher Scientific, GibcoTM, catalog number: 11965092 )
- BMDM Media (see Recipes)
- Culture Media (see Recipes)
- Treatment Media (see Recipes)
- FACS Buffer (see Recipes)
- L929-conditioned media (see Recipes)
- LPS EB-Ultrapure (lipopolysaccharide from E. coli O111:B4, InvivoGen, catalog number: tlrl-3pelps )
Equipment
- TC20 Automated cell counter (Bio-Rad Laboratories, catalog number: 1450102 )
- Pipet-aid Pipette Controller (Drummond Scientific, catalog number: 4-000-101 )
- 4 °C Cold Room
- 4 °C Benchtop centrifuge
- 37 °C Cell Culture Incubator with CO2 control
- Sterile cell culture hood
- Flow Cytometer (BD, model: FACSCantoTM II , or equivalent)
Software
- GraphPad Prism 7 (Graph Pad Software; La Jolla, CA USA)
Procedure
- Day 1, Cell culture
- Collect cultured bone marrow-derived macrophages (BMDMs) by gentle scraping. Disperse the cells into a single-cell suspension by repeatedly running the pipetted cell suspension along the test tube walls. Centrifuge cells (400 x g, 5 min, 23 °C), aspirate supernatant, and resuspend cell pellet in BMDM media. Count live cells via trypan blue exclusion assay.
- Plate 0.5 x 106 cells/well in a 6-well, non-treated, tissue culture plate. Culture the cells overnight in 2 ml/well of BMDM media. After 16 h of incubation (37 °C, 5% CO2), cells should be adherent and ready for the experiment.
- Collect cultured bone marrow-derived macrophages (BMDMs) by gentle scraping. Disperse the cells into a single-cell suspension by repeatedly running the pipetted cell suspension along the test tube walls. Centrifuge cells (400 x g, 5 min, 23 °C), aspirate supernatant, and resuspend cell pellet in BMDM media. Count live cells via trypan blue exclusion assay.
- Day 0, Preparation before LPS stimulation
- Prepare Culture and Treatment Media as described in Recipes. Warm the LPS-containing treatment media to 37 °C prior to use.
- Chill sterile PBS, 1.5 ml Eppendorf tubes, and FACS buffer to 4 °C, prior to use. After LPS treatment, the cells will be collected using these solutions and tubes.
- Prepare Culture and Treatment Media as described in Recipes. Warm the LPS-containing treatment media to 37 °C prior to use.
- Day 0, LPS stimulation of cells
- Aspirate BMDM media and wash 3 x with 3 ml of HBSS (room temperature) to remove dead cells and debris from each well. Add HBSS down the wall of the culture well and gently swirl plate to wash.
- Gently add 2 ml of Culture Media to wells labeled “Unstained BMDMs” and “t = 0 min/Untreated” treatment groups.
- Gently add 2 ml of Treatment Media to each well by pipetting the media along the side of the wells. Gently swirl the plate to ensure that the media is evenly distributed in the wells.
- Incubate at 37 °C for desired time points. Repeat Steps 3a to 3c as necessary for remaining LPS-treatment groups. Stagger the LPS treatment such that all samples are harvested at the same time.
- Aspirate BMDM media and wash 3 x with 3 ml of HBSS (room temperature) to remove dead cells and debris from each well. Add HBSS down the wall of the culture well and gently swirl plate to wash.
- Day 2, Cell collection and antibody staining
Note: All reagents should be cold and the procedure should be performed at 4 °C (cold room).- Transfer plates treated in Step 3 to 4 °C for 5 min prior to collection – this is required to arrest endocytosis.
- Aspirate media from each well. Wash 2 x each with 2 ml of sterile, pre-chilled PBS.
- Add 1 ml of sterile, pre-chilled PBS to each well. Gently scrape to detach cells and pipette-mix to disperse the cells into a single cell suspension.
- Transfer the cell suspension to 1.5 ml Eppendorf tubes and centrifuge (400 x g, 5 min) to pellet the cells.
- After aspirating and discarding the supernatant, resuspend the cell pellet in 50 μl of cold FACS Buffer premixed with TruStain fcXTM antibody (1 μg/ml), for 10 min.
- Add 50 μl of the 2x-concentrate of antibody (anti-TLR4 or anti-CD14; see Table 1) in FACS Buffer to the cell suspension. Add 50 μl of FACS Buffer to “Unstained BMDMs” samples.
Table 1. Antibodies used for measuring TLR4 and CD14 endocytosis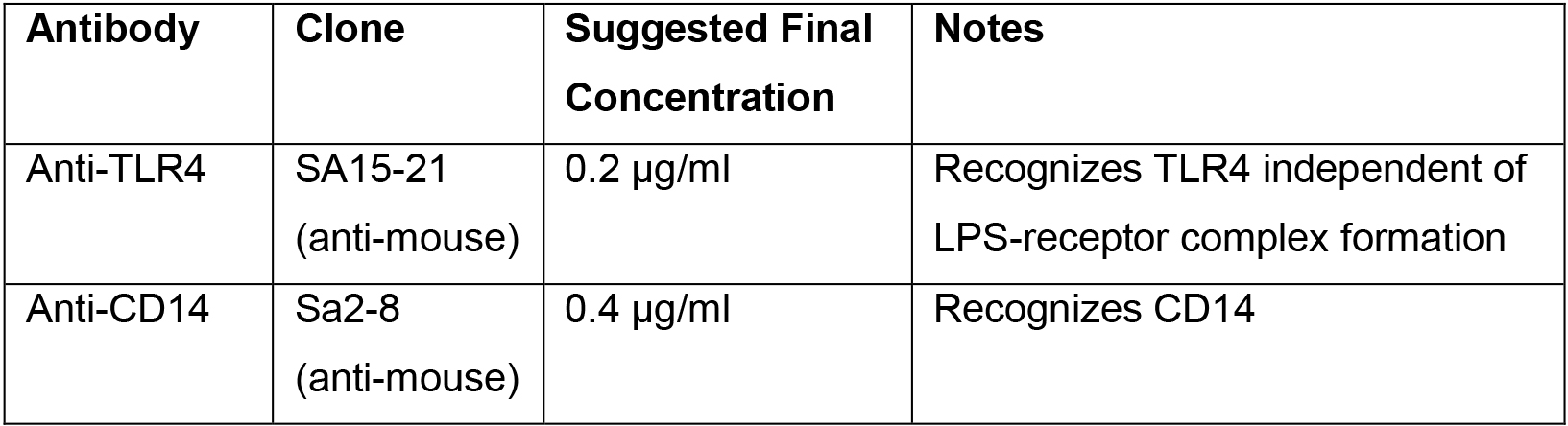
- Pipette gently to mix and stain for 20 min in the dark.
- Add 1 ml of FACS Buffer, collect cells by centrifugation (400 x g, 5 min) and aspirate supernatant to remove excess antibody.
- Resuspend the cell pellet in 200 μl of FACS Buffer and transfer the cell suspension to FACS tubes. Keep samples on ice and in the dark (e.g., cover with aluminum foil) prior to measurement by flow cytometry.
- Analyze samples via flow cytometry within 1 h.
- Transfer plates treated in Step 3 to 4 °C for 5 min prior to collection – this is required to arrest endocytosis.
Data analysis
- Flow cytometry analysis
- For flow cytometry analysis, collect > 100,000 events for each sample.
- For analysis, the events are gated on FSC-A and SSC-A bivariant cytographs; the low FSC-SSC events comprising of dead cells and cellular debris are excluded from analysis. Cells are then gated on FSC-A and FSC-H to gate on single cells.
- These cells can then be visualized for the intensity of the antibody stain as a histogram. Fluorescent intensity of the antibody stain on the gated population is recorded as the geometric mean of the cell population, or mean fluorescent intensity (MFI).
- For flow cytometry analysis, collect > 100,000 events for each sample.
- Data analysis for the measurement of TLR4 and CD14 Endocytosis
- For data analysis, the MFI of “Unstained cells” can be used for background subtraction from all samples [“Background Subtracted MFI”]. Divide the “background subtracted MFI” value for a given time point by the “Unstimulated [t = 0 min]” sample; “t = 0 min” value should be 1.00. Repeat this for all subsequent experimental samples to determine “Relative % of surface expression” relative to the “Unstimulated [t = 0 min]” sample. All sample values reflect the ratio of MFI from stimulated to unstimulated cells at desired time points.
- We convert the “Relative % of surface expression” value to a percentage; Thus, the “Untreated” or “Time = 0 min” sample should equal ‘100% percentage of surface expression’. One expects to see a steady reduction in this value at various time points after LPS stimulation.
- Since MFI values are sensitive to variations in flow cytometry calibrations, we recommend that data analysis be confined to each independent experiment and each condition run in technical triplicate. “Percentage of surface expression” should be reproducible across independent experiments and therefore amenable to statistical analysis of multiple experiments. Results from a typical experiment are shown in Figure 2. The original data presentation and additional information are available in our manuscript, which originally utilized the protocol described herein PubMed.
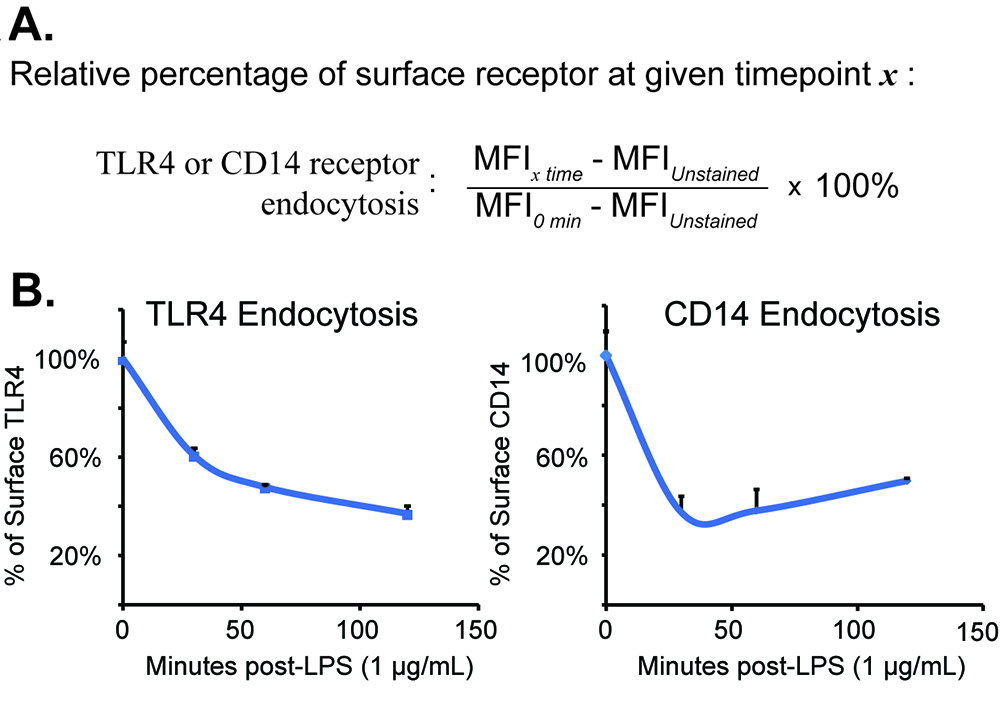
Figure 2. TLR4 and CD14 receptor endocytosis data analysis and suggested presentation. A. “Data analysis” calculation described as a formula. B. Characteristic TLR4 and CD14 endocytosis measured over time in bone-marrow derived macrophages. Data was modified from its original presentation in Schappe et al., 2018 with author permission.
- For data analysis, the MFI of “Unstained cells” can be used for background subtraction from all samples [“Background Subtracted MFI”]. Divide the “background subtracted MFI” value for a given time point by the “Unstimulated [t = 0 min]” sample; “t = 0 min” value should be 1.00. Repeat this for all subsequent experimental samples to determine “Relative % of surface expression” relative to the “Unstimulated [t = 0 min]” sample. All sample values reflect the ratio of MFI from stimulated to unstimulated cells at desired time points.
- For experimental and statistical analysis, we use GraphPad Prism. To compare two experimental groups, we use a Student’s t-test. For comparison of three or more data groups, other statistical analysis, such as a one-way ANOVA, are necessary.
Notes
- The murine macrophage RAW 264.7 cell line also exhibits characteristic TLR4 endocytosis–it can be used to establish the method and for experiments.
- Although non-treated culture plates may permit detachment with trypsin, the enzymatic detachment may alter macrophage epitope expression at the plasma membrane.
- Performing “Procedure” Step 4 in a walk-in 4 °C cold room greatly improves the quality of data. Although chilling materials and reagents on ice may be convenient, variations in temperature between the ice, samples, and ambient laboratory air may inadvertently warm samples above 4 °C, thereby permitting endocytosis to proceed.
- Although procedural steps after “Procedure” Step 3d are not performed under sterile conditions, using sterile reagents minimizes inadvertent contamination with ligands that may promote TLR4 or CD14 endocytosis.
- Although spectrally non-overlapping fluorophores are available, we advise staining with a single anti-TLR4 or CD14 antibody for each experiment.
- With careful spectral consideration, fluorescent live/dead dyes can be included in this assay to enrich for live cell populations. If the experimenter includes these dyes, we advise that only spectrally-compatible nucleic acid binding dyes, which can rapidly label dead cells during the final suspension be used. Some viability dyes and staining methods (such as “live/dead fixable dyes” or Annexin V-based staining kits) require additional staining steps that may compromise the time and temperature-sensitivity of this assay.
- Titration of antibodies, including new batches of the same antibody clone, is essential. Although recommendations are provided in Table 1, improper staining will limit signal-to-noise ratio (SNR) in the assay and lower data quality in terms of sensitivity and consistency.
- Avoid sample groups larger than 24 samples to minimize sample processing time prior to flow cytometry analysis.
- The majority of our data were collected on the BD FACSCanto II flow cytometer.
Recipes
- BMDM Media
- RPMI 1640 + 10% FBS + 20% L929-conditioned media
- Store at 4 °C for up to 1 month
- RPMI 1640 + 10% FBS + 20% L929-conditioned media
- Culture Media
- RPMI 1640 + 10% FBS
- Store at 4 °C for up to 1 month
- RPMI 1640 + 10% FBS
- Treatment Media
- RPMI 1640 + 10% FBS + 1 μg/ml LPS
- Prepare fresh for each experiment
- RPMI 1640 + 10% FBS + 1 μg/ml LPS
- FACS Buffer
- PBS + 1% BSA
- Sterile filter buffer through 0.22 μm filter prior to use
- Store at 4 °C for up to 1 week
- PBS + 1% BSA
- L929-conditioned Media
Note: Generated from the culture of L-929 cells (available from ATCC). Cells are passaged according to the vendor’s instructions and cultured in DMEM, high glucose + 10% FBS.- To generate L929-conditioned media, add 150 ml of DMEM, high glucose + 10% FBS to a T-150 TripleFlask. Add 0.72 x 106 L-929 cells and carefully mix by equilibrating the media volume at the hole in the corner of the flask. Culture for 7 days at 37 °C, 5% CO2
- On Day 7, collect media, sterile filter through a 0.22 μm filter into a flask, and store at -20 °C [“Week 1 media”]; add 150 ml of DMEM, high glucose + 10% FBS to TripleFlask to replace collected media
- On Day 14, collect media from the flask and sterile filter through a 0.22 μm filter [“Week 2 media”]. Thaw Week 1 media at 23 °C
- Combine Week 1 and Week 2 media and aliquot into 50 ml tubes. L929-conditioned media is stored at -20 °C for up to 6 months
- To generate L929-conditioned media, add 150 ml of DMEM, high glucose + 10% FBS to a T-150 TripleFlask. Add 0.72 x 106 L-929 cells and carefully mix by equilibrating the media volume at the hole in the corner of the flask. Culture for 7 days at 37 °C, 5% CO2
Acknowledgments
We would like to thank Dr. Jonathan Kagan (Boston Children’s Hospital) and colleagues whose protocol (cited herein) we adapted for our research. We also would like to thank members of the Desai laboratory for their assistance in editing this manuscript. We also thank the UVA Flow Cytometry Core and Carter Immunology Center Flow Cytometry Core. We appreciate our funding that supported this work: grants GM108989 (BND) and 5T32GM007055-41 (MSS) from the National Institutes of Health.
The authors declare that they have no conflicts of interest to report.
References
- Brubaker, S. W., Bonham, K. S., Zanoni, I. and Kagan, J. C. (2015). Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33: 257-290.
- Kagan, J. C., Su, T., Horng, T., Chow, A., Akira, S. and Medzhitov, R. (2008). TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat Immunol 9(4): 361-368.
- Roy, S., Karmakar, M. and Pearlman, E. (2014). CD14 mediates Toll-like receptor 4 (TLR4) endocytosis and spleen tyrosine kinase (Syk) and interferon regulatory transcription factor 3 (IRF3) activation in epithelial cells and impairs neutrophil infiltration and Pseudomonas aeruginosa killing in vivo. J Biol Chem 289(2): 1174-1182.
- Schappe, M. S., Szteyn, K., Stremska, M. E., Mendu, S. K., Downs, T. K., Seegren, P. V., Mahoney, M. A., Dixit, S., Krupa, J. K., Stipes, E. J., Rogers, J. S., Adamson, S. E., Leitinger, N. and Desai, B. N. (2018). Chanzyme TRPM7 mediates the Ca2+ influx essential for lipopolysaccharide-induced toll-like receptor 4 endocytosis and macrophage activation. Immunity 48(1): 59-74 e55.
- Tan, Y., Zanoni, I., Cullen, T. W., Goodman, A. L. and Kagan, J. C. (2015). Mechanisms of toll-like receptor 4 endocytosis reveal a common immune-evasion strategy used by pathogenic and commensal bacteria. Immunity 43(5): 909-922.
- Zanoni, I., Ostuni, R., Marek, L. R., Barresi, S., Barbalat, R., Barton, G. M., Granucci, F. and Kagan, J. C. (2011). CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147(4): 868-880.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Schappe, M. S. and Desai, B. N. (2018). Measurement of TLR4 and CD14 Receptor Endocytosis Using Flow Cytometry. Bio-protocol 8(14): e2926. DOI: 10.21769/BioProtoc.2926.
Category
Immunology > Immune cell function > Macrophage
Cell Biology > Cell-based analysis > Cytosis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link