- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Induction of Natural Competence in Genetically-modified Lactococcus lactis
Published: Vol 8, Iss 13, Jul 5, 2018 DOI: 10.21769/BioProtoc.2922 Views: 7766
Reviewed by: Modesto Redrejo-RodriguezAdison WongAlba Blesa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Fast and Easy Method to Study Ralstonia solanacearum Virulence upon Transient Gene Expression or Gene Silencing in Nicotiana benthamiana Leaves
Wenjia Yu and Alberto P. Macho
Aug 5, 2021 4699 Views

A New Tool for the Flexible Genetic Manipulation of Geobacillus kaustophilus
Ryotaro Amatsu [...] Ken-ichi Yoshida
Sep 5, 2022 1905 Views

Efficient Genetic Transformation and Suicide Plasmid-mediated Genome Editing System for Non-model Microorganism Erwinia persicina
Tingfeng Cheng [...] Lei Zhao
Mar 20, 2024 2430 Views
Abstract
Natural competence can be activated in Lactoccocus lactis subsp lactis and cremoris upon overexpression of ComX, a master regulator of bacterial competence. Herein, we demonstrate a method to activate bacterial competence by regulating the expression of the comX gene by using a nisin-inducible promoter in an L. lactis strain harboring either a chromosomal or plasmid-encoded copy of nisRK. Addition of moderate concentrations of the inducer nisin resulted in concomitant moderate levels of ComX, which led to an optimal transformation rate (1.0 x 10-6 transformants/total cell number/g plasmid DNA). Here, a detailed description of the optimized protocol for competence induction is presented.
Keywords: Natural competenceBackground
Natural competence is the process in which a bacterium acquires exogenous DNA via a specialized uptake machinery after which the internalized DNA is either integrated into its genome or maintained as plasmid DNA. Several bacteria enter a state of competence upon specific environmental triggers such as genotoxic stress or starvation (Seitz and Blokesch, 2013; Blokesch, 2016). Quorum sensing systems such as comCDE or comRS control the activation of natural competence in Gram positive bacteria (Håvarstein et al., 1995; Pestova et al., 1996; Kleerebezem et al., 1997b; Fontaine et al., 2015). More specifically, comC and comS encode pheromones, whereas comD encodes a histidine kinase and comE and comR encode response regulators (Håvarstein et al., 1995; Pestova et al., 1996; Fontaine et al., 2010; Fontaine et al., 2015). In streptococci, the activated regulator drives transcription of the alternative sigma factor ComX, which in turn, activates transcription of competence genes that encode the proteins encompassing the DNA uptake machinery (Johnston et al., 2014). Previously, strategies employing overexpression of ComX led to the successful introduction of exogenous DNA in Streptococcus thermophilus (Blomqvist et al., 2006) and L. lactis (David et al., 2017; Mulder et al., 2017), even though different approaches were employed to achieve its overexpression. These studies also showed that different expression levels of comX critically impact on transformation rates. For example, in our work we used L. lactis subsp. lactis KF147, a strain that allows Nisin-Controlled gene Expression system (NICE) (Mierau and Kleerebezem, 2005) and harbors chromosomal nisRK (essential to allow nisin induction) but does not produce nisin. In this strain, we introduced pNZ6200, a vector containing comX under the control of the nisin-inducible nisA promotor, by electro-transformation, and the resulting strain was not transformable upon full comX induction (2 ng/ml nisin), whereas optimal transformation rates (1.0 x 10-6 transformants/total cell number/g plasmid DNA) were observed upon moderate levels of induction (0.03 ng/ml nisin) (Mulder et al., 2017). Moreover, we also demonstrated that applying the same strategy worked for a nisRK derivative of L. lactis subsp. lactis IL1403, whereas competence induction in L. lactis subsp. cremoris KW2 required prior transformation of the strain with pNZ9531 that expresses nisRK (Kleerebezem et al., 1997a) to allow nisin induced expression of comX. In order to assess whether other L. lactis strains can become naturally competent, we hereby provide a method containing all details of the competence protocol as described previously (Mulder et al., 2017) that can assist other scientists to unleash competence in the L. lactis strain of their interest and might prevent experimental issues concerning nisin induced expression of ComX. This newly developed protocol is expected to allow genetic access in a broad panel of L. lactis strains with an efficiency that enables rapid-one-step construction of gene replacement mutants via integration of linear DNA fragments harboring an antibiotic resistance marker flanked by chromosomal homologous DNA regions.
Materials and Reagents
- Pipette tips
- 15 ml and 50 ml CELLSTAR® Polypropylene Tube (conical) (Greiner Bio One International, CELLSTAR®, catalog numbers: 188271 and 227261 respectively)
- Plastic Petri dish 94 x 16 with vents light version (Greiner Bio One International, catalog number: 633181 )
- 12 ml Cell Culture Tubes (Greiner Bio One International, CELLSTAR®, catalog number: 163160 )
- Inoculation loops
- Eppendorf Tubes® Safe-Lock Tubes 1.5 ml (Eppendorf, catalog number: 0030120086 )
- Electroporation cuvettes, 2 mm gap (Bio-Rad Laboratories, catalog number: 1652086 )
- Semi-micro cuvettes for 1 ml (for optical density measurements)
- Bacterial strains: L. lactis strain of interest with a complete set of competence genes, L. lactis harboring pNZ6200 (Mulder et al., 2017), pNZ6202 (Mulder et al., 2017), and pNZ9531 (Kleerebezem et al., 1997a)
- M17 broth (M17) and M17 agar (M17A) (Tritium, catalog numbers: M086.76.0200 and M085.76.0200 respectively)
- Glucose solution (20%, Tritium, catalog number: G209.65.0080 )
- Distilled water (Thermo Fisher Scientific, GibcoTM, catalog number: 15230089 )
- Stock solutions of antibiotics:
20 mg/ml chloramphenicol (Sigma-Aldrich, catalog number: C0378-100G )
20 mg/ml erythromycin (Fisher Scientific, catalog number: BP920-25 )
12.5 mg/ml Tetracycline hydrochloride (Sigma-Aldrich, catalog number: T7660-25G ) - NisinA® P Ultrapure Nisin A (Handary, Brussels, Belgium, prepare a 2 mg/ml stock solution in distilled water containing 0.05 glacial acetic acid)
- Alternatively: nisin from Lactococcus lactis 2.5% (Sigma-Aldrich, catalog number: N5764 )
- JETSTAR 2.0 Maxiprep Kit (GENPRICE, catalog number: 220 020 ) or PureLinkTM HiPure Plasmid Maxiprep Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: K210007 )
- QubitTM dsDNA BR Assay Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32850 )
- Phenol BioUltra, for molecular biology, TE-saturated, ~73% (T) (Sigma-Aldrich, catalog number: 77607 )
- Chloroform HPLC grade, ≥ 99.9% (Sigma-Aldrich, catalog number: 528730 )
- Ethidium bromide (Sigma-Aldrich, catalog number: E1510-10ML )
- Agarose tablets (U.S. Biotech Sources, catalog number: G01PD-500 )
- Glacial acetic acid (Scharlab, catalog number: AC03522500 )
- β-glycerophosphate (Disodium salt) (Sigma-Aldrich, catalog number: 50020-500G )
- Potassium phosphate dibasic (K2HPO4) (Merck, catalog number: 1.05104.1000 )
- Potassium phosphate monobasic (KH2PO4) (Merck, catalog number: 1.04873.1000 )
- Na-acetate (Merck, catalog number: 1.06268.1000 )
- (NH4)3-citrate (Sigma-Aldrich, catalog number: A1332-500G )
- Ascorbic acid (VWR, AnalaR NORMPAPUR®, catalog number: 20150.231 )
- Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Sigma-Aldrich, catalog number: M8054-100G )
- Adenine (Sigma-Aldrich, Fluka, catalog number: 01830 )
- Guanine (Sigma-Aldrich, catalog number: G11950-100G )
- Uracil (Sigma-Aldrich, Fluka, catalog number: 94220 )
- Xanthine (Sigma-Aldrich, catalog number: X7375-10G )
- Alanine (Sigma-Aldrich, catalog number: A7627-100G )
- Arginine (Sigma-Aldrich, catalog number: A5006-500G )
- Aspartic acid (Sigma-Aldrich, catalog number: A9256-100G )
- Cysteine-HCl (Sigma-Aldrich, catalog number: C1276-250G )
- Glutamic acid (Sigma-Aldrich, catalog number: G1251-500G )
- Glycine (Merck, catalog number: 1.04201.0250 )
- Histidine (Sigma-Aldrich, catalog number: H8000-100G )
- Leucine (Sigma-Aldrich, catalog number: L8000-100G )
- Lysine (Sigma-Aldrich, catalog number: L5626-100G )
- Methionine (Sigma-Aldrich, catalog number: M9625-100G )
- Phenylalanine (Sigma-Aldrich, catalog number: P2126-100G )
- Proline (Sigma-Aldrich, catalog number: P0380-100G )
- Serine (Sigma-Aldrich, catalog number: S4500-100G )
- Threonine (Sigma-Aldrich, catalog number: T8625-100G )
- Tryptophane (Sigma-Aldrich, catalog number: T0254-100G )
- Valine (Sigma-Aldrich, catalog number: V0500-100G )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M2670-100G )
- Calcium chloride dihydrate (CaCl2·2H2O) (Merck, catalog number: 1.02382.0500 )
- Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sigma-Aldrich, catalog number: Z0251-100G )
- Cobalt(II) sulfate heptahydrate (CoSO4·7H2O) (Sigma-Aldrich, catalog number: C6768-100G )
- Copper (II) sulfate pentahydrate (CuSO4·5H2O) (Scharlab, catalog number: CO01010500 )
- Ammonium molybdate tetrahydrate ((NH4)6Mo7O24·4H2O) (Sigma-Aldrich, Fluka, catalog number: 09878 )
- Iron(II) chloride tetrahydrate (FeCl2·4H2O) (Sigma-Aldrich, catalog number: 44939-50G )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 30721-1L )
- Iron(III) chloride hexahydrate (FeCl3·6H2O) (Sigma-Aldrich, catalog number: F2877-100G )
- p-aminobenzoëic acid (Sigma-Aldrich, catalog number: A9879-100G )
- Inosine (Sigma-Aldrich, catalog number: I4125-25G )
- Orotic acid (Sigma-Aldrich, catalog number: O2750-100G )
- Pyridoxamine-HCl (Sigma-Aldrich, catalog number: P9380-5G )
- Thymidine (Sigma-Aldrich, catalog number: T9250-10G )
- D-biotin (Sigma-Aldrich, catalog number: B4501-5G )
- 6,8-thioctic acid (Sigma-Aldrich, catalog number: T5625-5G )
- Pyridoxine-HCl (Sigma-Aldrich, catalog number: P9755-25G )
- Folic acid (Sigma-Aldrich, catalog number: F7876-25G )
- Nicotinic acid (Sigma-Aldrich, catalog number: N4126-5G )
- Ca-(D+)pantothenate (Sigma-Aldrich, catalog number: P5155-100G )
- Riboflavin (Sigma-Aldrich, catalog number: R4500-25G )
- Thiamin-HCl (Sigma-Aldrich, catalog number: T4625-100G )
- Vitamin B12 (Sigma-Aldrich, catalog number: V2876-5G )
- Alternatively, PCR product (obtained with KOD hot start DNA polymerase)
Note: Containing an antibiotic resistance marker flanked by chromosomal fragments for integration obtained by using overlap PCR (Horton et al., 1990) if preferred over plasmid transformation, e.g., for the construction of gene replacement mutants. - Optional: KOD hot start DNA polymerase Master Mix (Merck, Novagen, catalog number: 71842-4 )
- GSGM17 (see Recipes)
- Washing solution 1 (see Recipes)
- Washing solution 2 (see Recipes)
- Recovery medium (see Recipes)
- CDM (see Recipes)
- GCDM (chemically defined medium supplemented with glucose) (see Recipes)
- Stock solutions for CDM (see Recipes)
- Nucleotide solution
- MnCl2·4H2O solution
- Amino acid solution
- Metal solution
- Iron solution
- Vitamin solution
- Nucleotide solution
Equipment
- General pipettes
- 200 ml bottle
- Water bath (for 30 °C and 55 °C)
- Incubation stove at 30 °C
- Vortex
- Autoclave
- Microcentrifuge (Eppendorf® Refrigerated Microcentrifuge) (Eppendorf, model: 5417R )
- Centrifuge for 15 and 50 ml tubes (Heraeus Megafuge 1.0R)
- Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, InvitrogenTM, mode: Qubit® 2.0 )
- Electroporator (Bio-Rad Laboratories, model: GenePulserXcellTM )
- Spectrophotometer (Genesys 10 UV Spectrophotometer)
- Nanodrop ND1000 spectrophotometer (NanoDrop Technologies) (Thermo Fisher Scientific, model: NanoDropTM 1000 )
Procedure
- Plasmid DNA isolation
- Culture L. lactis harboring either pNZ6200, pNZ6202, or pNZ9531 in 200 ml M17 supplemented with 1% glucose in a 200 ml bottle containing the appropriate antibiotic (Table 1) at 30 °C without shaking overnight.
Table 1. Characteristics of plasmids used in this protocol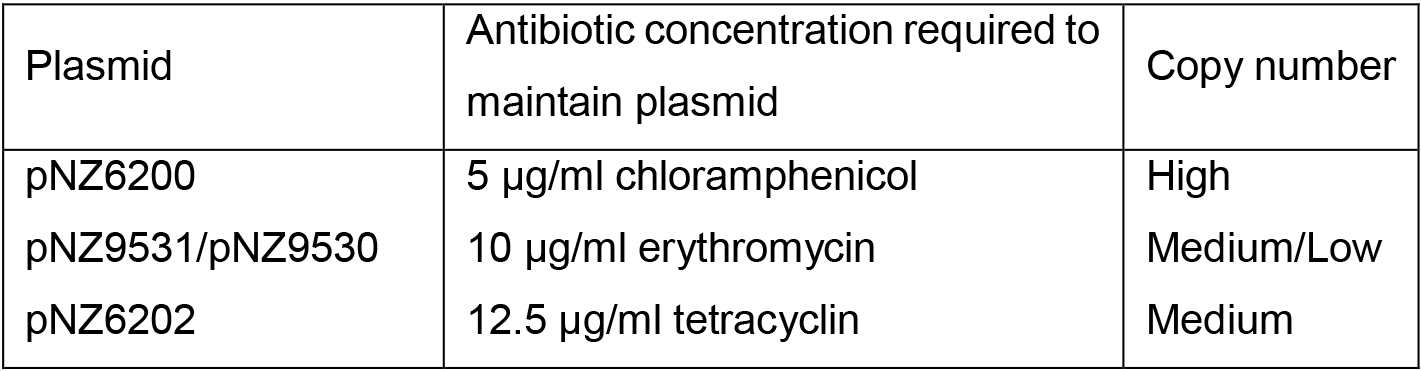
- Pellet cells by centrifugation at 5,000 x g for 15 min at room temperature.
- Discard the culture supernatant and freeze the pellets at -20 °C for at least 30 min.
- Resuspend the pellets in 10 ml resuspension buffer (manufacturer’s solution from the Maxiprep Kit) containing 40 mg lysozyme.
- Incubate for 1.5 h at 55 °C without shaking.
- Proceed with the plasmid isolation according to the manufacturer’s protocol from either the JETSTAR 2.0 Maxiprep Kit or PureLinkTM HiPure Plasmid Maxiprep Kit.
- Include some extra steps to the manufacturer’s protocol: After centrifugation of the lysates to pellet cell debris, perform phenol-chloroform extraction on the supernatants (phenol: chloroform: supernatant in 1:1:1 volume ratio, mix by inverting the tube) and centrifuge at 5,000 x g for 30 min. Transfer the aqueous phase (upper layer) to a new tube and subject to chloroform extraction (aqueous phase: chloroform 1:1 volume) and subsequent centrifuge at 5,000 x g for 15 min. Load purified fractions on the column from the manufacturer’s kit to allow binding of the DNA and continue the steps as indicated in the manual supplied.
- Determine DNA concentration of plasmid DNA by Qubit (1 µg of DNA is required per transformation where the concentration of the plasmid solution is based on Qubit measurement).
- Check the quality of the isolated plasmid DNA on a 1% agarose gel.
- Culture L. lactis harboring either pNZ6200, pNZ6202, or pNZ9531 in 200 ml M17 supplemented with 1% glucose in a 200 ml bottle containing the appropriate antibiotic (Table 1) at 30 °C without shaking overnight.
- Electrotransformation of pNZ6200 and pNZ9531 into electrocompetent L. lactis cells (Figure 1)
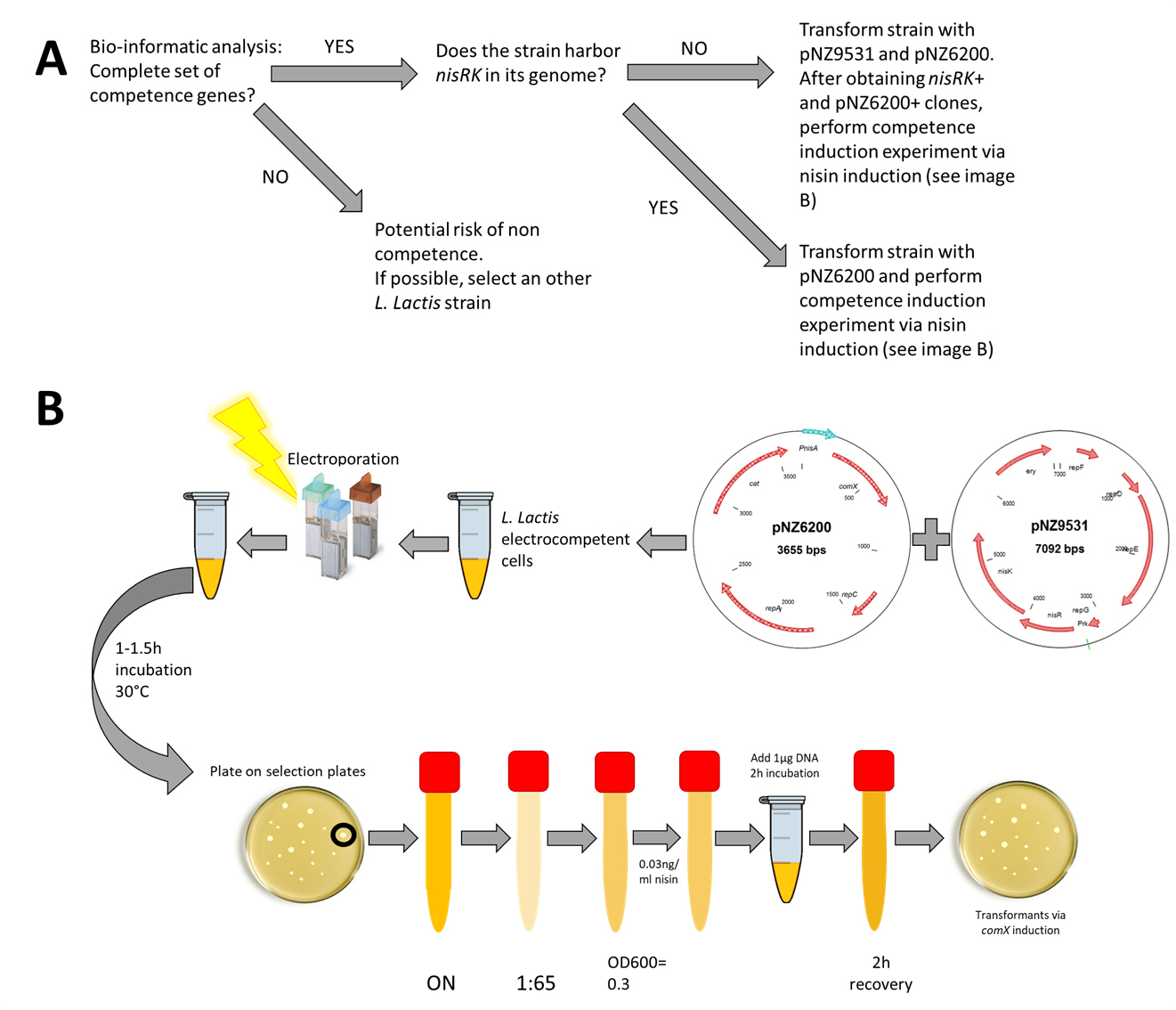
Figure 1. Schematic overview for evaluation of competence potential (A) and obtaining an L. lactis strain harboring pNZ6200 and pNZ9531, as well as subsequent induction of competence via nisin induction (B). See the notes in the procedure section for specific cases where we advise modifications to this standard flow scheme. - Prepare 1 L of the electro competence growth medium GSGM17, 500 ml washing solution 1 and 500 ml washing solution 2 (see Recipes).
- Culture L. lactis initially overnight in 10 ml GM17 in a 12 ml Cell Culture Tube, incubate at 30 °C without shaking.
- The next day, inoculate 5 ml culture into 50 ml GSGM17 and incubate cells overnight at 30 °C, without shaking.
- Dilute the overnight culture 1:8 in GSGM17 if the overnight culture reached an OD600 of ≥ 0.7 and grow the culture to an OD600 of 0.2-0.3.
- Pellet cells by centrifugation at 6,000 x g for 20 min at 4 °C.
- Wash the pellet with 1 volume ice-cold washing solution 1.
- Pellet cells by centrifugation at 5,000 x g for 20 min at 4 °C.
- Wash the pellet with 0.5 volume ice-cold washing solution 2 and incubate on ice for 15 min.
- Pellet cells by centrifugation at 5,000 x g for 20 min at 4 °C.
- Wash the pellet with 0.25 volume ice-cold washing solution 1 and pellet cells by centrifugation at 5,000 x g for 20 min at 4 °C.
- Resuspend cells in 0.01 volume ice-cold washing solution 1 and aliquot cells per 50 µl.
- Use fresh electrocompetent cells immediately for transformation with the desired plasmid.
Option: Store cells for later use at -80 °C. - Keep electro-cuvettes on ice and prepare recovery medium containing M17 supplemented with 1% glucose, 200 mM MgCl2 and 20 mM CaCl2 and store it on ice as well.
- Add 1 µg plasmid to electrocompetent cells and transfer the cell suspension with DNA into the electro-cuvette.
Note: Do not pipet up and down after addition of plasmid DNA to the electrocompetent cells. Mix cells and plasmid DNA by gently flicking the tube. - Electroporate; settings for the electroporator device: voltage = 2,000 V, capacity = 25 µF, resistance = 200 Ω.
- Dry the electro-cuvette with a tissue and tap the electro-cuvette gently onto the bench 5 times to slip down the cells to the bottom of the electro-cuvette.
- Put the electro-cuvette in the cuvette holder of the electroporator and apply the electrical pulse.
- Dry the electro-cuvette with a tissue and tap the electro-cuvette gently onto the bench 5 times to slip down the cells to the bottom of the electro-cuvette.
- Add 1 ml of ice-cold recovery medium as soon as possible to the electro-cuvette and put the electro-cuvette on ice again.
- Pipet the cells from the electro-cuvette into a 1.5 ml Eppendorf tube and incubate samples in a 30 °C water bath for 1 h without shaking.
- Pipet serial dilutions for 100 to 10-6 in recovery medium on selection plates with appropriate antibiotics.
- Incubate plates for 2-3 days at 30 °C under non-shaking conditions.
- Clean streak positive colonies on a GM17 agar plate supplemented with appropriate antibiotics.
Note: If co-transformation of pNZ9531 and pNZ6200 is unsuccessful, transform cells first with pNZ9531. Select positive colonies and culture colonies in GM17 supplemented with 10 µg/ml erythromycin. Subsequently, repeat the preparation for electro competence cells and introduce pNZ6200 by electroporation. - The next day, culture single colonies from the clean streak plate in GM17 supplemented with appropriate antibiotics and incubate overnight at 30 °C under non-shaking conditions.
- Prepare a 25% glycerol stock and store at -80 °C.
- Induction of natural competence in L. lactis
- Prepare GCDM (see Recipes) and prepare GM17 agar plates with appropriate antibiotics.
- Culture L. lactis harboring pNZ6200 and pNZ9531 separately in 10 ml GCDM supplemented with the appropriate antibiotics (Table 1).
- Prepare a 50 ml CELLSTAR® Polypropylene Tube containing 50 ml GCDM.
- Incubate cells and the 50 ml CELLSTAR® Polypropylene Tube containing 50 ml GCDM at 30 °C overnight in an incubation stove without shaking.
- The next day, add chloramphenicol and erythromycin to the 50 ml CELLSTAR® Polypropylene Tube with 50 ml GCDM.
- Add 1 ml of the GCDM in a semi-micro cuvette (= blank sample) by measuring optical density at OD600.
- Add 750 µl of the L. lactis harboring pNZ6200 overnight culture to the 50 ml CELLSTAR® Polypropylene Tube tube containing GCDM.
- Mix the cell culture by inverting the closed tube.
- Pipet 1 ml of the culture into a semi-micro cuvette and incubate cells in the 30 °C water bath non-shaking until the culture reaches an OD600 of 0.3 (after approximately 3-4 h).
- Measure the initial OD600 of the culture.
Note: The OD600 of the culture at t = 0 should be around 0.03. If the OD600 is lower than 0.03; add more culture. - Culture cells until an OD600 of 0.3 (corresponds with approximately 2-3 x 108 cells) under non-shaking conditions.
- Prepare calculations and recovery medium (see Recipes, 5 ml recovery medium per transformation).
- Transfer 10 ml of cell culture at OD600 of 0.3 into a 12 ml sterile tube.
- Repeat this step to obtain a total of 3 aliquoted cultures.
Note: These cultures will be used for the later steps involving the induction of comX expression with 0 ng/ml (negative control), 0.03 ng/ml (optimal) and 2 ng/ml (fully induced) nisin. - Subsequently, prepare the fresh nisin dilutions by using either NisinA® P Ultrapure Nisin A or nisin from Lactococcus lactis 2.5% (Sigma-Aldrich, 2.5% nisin).
- Prepare nisin dilutions in glass-tubes by using distilled water containing 0.05% glacial acetic acid in order to increase the stability of the nisin solutions.
Note: When using nisin from L. lactis 2.5% (Sigma-Aldrich), make sure you correct for the dilution factor (40x) as only 2.5% of the dry matter is nisin. - Induce the 10 ml culture with nisin. For optimal transformation rates, induce cells with nisin to a final concentration of 0.03 ng/ml nisin.
Notes:- Optional: as a control, include an uninduced sample and a fully induced sample (2 ng/ml).
- L. lactis KF147 harboring pNZ6200 induced with 2 ng/ml nisin will stop growth after 1 h induction but other strains might continue growth at this concentration.
- Optional: as a control, include an uninduced sample and a fully induced sample (2 ng/ml).
- Mix immediately by inverting the tubes 3 times after addition of nisin.
- Put 600 µl of the induced culture in an Eppendorf tube.
- Add 1 µg pNZ6202.
Note: It is likely that other compatible plasmids can also be used to assess transformation efficiencies. - Mix by inverting the tube.
- Incubate at 30 °C for 2 h in a water bath without shaking.
Note: Longer incubation with nisin does not lead to increased transformation rates (see Figure 2).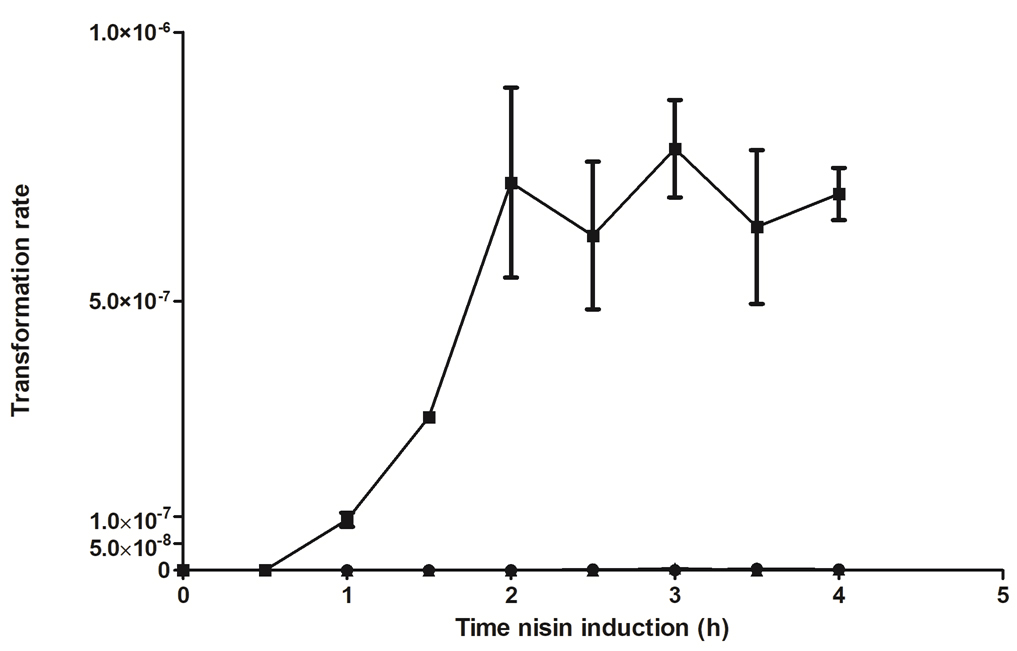
Figure 2. Transformation rate (transformants/total cell number after induction/µg plasmid DNA) after prolonged nisin induction in L. lactis KF147 harboring pNZ6200. Transformation was examined in uninduced (closed circles), moderately induced (closed squares) and fully induced (closed triangles) cultures. Uninduced and fully induced L. lactis KF147 harboring pNZ6200 were not transformable after shortened, standard (2 h) and prolonged induction with nisin. Moderately induced L. lactis KF147 harboring pNZ6200 could be transformed after prolonged nisin induction, however, prolonged induction did not lead to increased transformation rates as compared with the standard incubation time. - Pipet 100 µl of each sample into a semi-micro cuvette containing 900 µl GM17 and measure the OD600 of the sample or perform a total cell count.
Note: Take 1 ml of GM17 as a new blank. - Add the remaining 500 µl in the 5 ml recovery medium in a 15 ml tube and incubate for another 2 h at 30 °C in a water bath non-shaking.
Note: When transforming a plasmid or PCR product that contains a resistance gene against bacteriostatic antibiotics, this recovery step is not necessary. - Add 250 µl of the culture in a semi-micro cuvette containing 750 µl GM17 to measure OD600 or perform a total cell count.
- Pellet the rest of the culture in the 15 ml CELLSTAR® Polypropylene Tube by centrifugation at 4,000 x g for 10 min.
- Discard supernatant, leave 100 µl medium onto the pellet, resuspend and plate all cells on GM17A (M17 agar supplemented with 2% glucose) with appropriate antibiotics.
- Keep the plates in the incubation stove at 30 °C for 2 to 3 days.
- Calculate the transformation rate as (transformants/total cell number after induction/µg plasmid DNA).
- Sequence similarity of competence proteins from the strain of interest to query sequences BlastP analysis should be performed in order to assess the completeness of the competence system in the strain of interest by comparing its competence protein sequences to the query sequences from either L. lactis subsp. lactis KF147 or L. lactis subsp. cremoris KW2 (Appendix 1; 2; 3). As previously described, a virtually full-length alignment (> 90%) of the subject sequence to the query sequence is considered indicative of gene presence (Mulder et al., 2017). Commonly, e-values of 10-5 indicate significant alignments. If desired, genetic events leading to decay of competence genes can be further analyzed by bioinformatics software such as Clone Manager suite.
- Calculation of transformation rates
At least three replicates should be included in each experiment in order to calculate the average transformation rate of a nisin-induced L. lactis strain. Transformation rates can be shown either in tables or (in case of multiple time points) in a graph as depicted in Figure 1 by using Graphpad Prism or any other software. - Preparation of electrocompetent cells of L. lactis was based on Wells et al., 1993.
- If a strain displays poor growth in the glycine-containing medium (i.e., an overnight culture does not reach an OD600 of 0.7), the strain requires adaptation to GSGM17 medium by diluting this medium with regular GM17 (M17 supplemented with 2% glucose) 1:4, followed by serial subculturing in media with increasing amounts of glycine, to eventually reach the 0.4 M glycine concentration in GSGM17. If cells fail to adapt to the presence of 0.4 M glycine in GSGM17, then use the highest concentration of glycine possible.
- The L. lactis strain of interest should harbor a complete set of competence genes to allow competence induction upon comX overexpression. Completeness of the com gene set can be assessed by BlastP analysis using reference sequences for either subsp. lactis (Appendix 1) or subsp. cremoris as query (Appendix 2).
- The analogous alternative plasmid for pNZ9531, pNZ9530, can likely also be used to obtain a nisRK+ strain. In fact, it is expected that a tighter control of nisRK expression can be achieved when using pNZ9530 (Kleerebezem et al., 1997a).
- For extraction of pNZ9530 or pNZ9531 (low and medium copy plasmids, respectively) from an L. lactis strain: inoculate this strain in 25 ml GM17 supplemented with 10 µg/ml erythromycin without agitation. The next day, prepare 4 bottles of 200 ml GM17 supplemented with 10 µg/ml erythromycin and transfer 5 ml overnight culture to each bottle. Culture cells to an OD600 of 0.5-1 and pellet cells by centrifugation at 5,000 x g for 15 min. Proceed with Step 3 from the Plasmid DNA Isolation procedure.
- Presence of a chromosomal copy of nisRK in the L. lactis strain of interest can be examined (use Appendix 3 for query sequences of NisR and NisK for a BlastP analysis). If the strain of interest harbors nisRK, pNZ9530/pNZ9531 and subsequent transformation with one of these plasmids is not necessary.
- However, if the strain harbors nisRK which is also a nisin producer, this might result in constitutive induction of comX which may not lead to the favorable conditions for competence induction. In this case, an alternative could be transforming cells with pGIBLD001 (constitutive expression of comX under P32 control [David et al., 2017]).
- If for whatever reason GCDM cannot be used as culturing medium for L. lactis competence, M17 can be used, however, transformation rates are reduced by at least 10 fold for L. lactis KF147 harboring pNZ6200 (data not shown). If competence induction needs to be performed in medium that does not contain fluorescent components such as riboflavin, we recommend using CDMPC (chemically defined medium prolonged cultivation [Goel et al., 2012]) though transformation rates are reduced (1 x 10-7 transformants/total cell number after induction/µg plasmid DNA) for L. lactis KF147 harboring pNZ6200 in this medium.
- Induction of competence in L. lactis can also be performed in a high-throughput setup. However, we recommend inducing the culture in the 12 ml Cell Culture Tubes first, collect a 600 µl sample of the induced culture to add 1 µg DNA and seed 3 times 200 µl each time into a 96 wells plate. After 3 h of induction, 5 µl can be used for spot plating on GM17 plates with appropriate antibiotics by using a multichannel pipet. Centrifuge the rest of the culture in a plate centrifuge for 10 min at 3,700 x g and discard 150 µl supernatant. Resuspend the pelleted cells in the leftover medium and spot with a multichannel pipet 5 µl on a GM17 plate with appropriate antibiotics. For L. lactis KF147 harboring pNZ6200, we were able to obtain similar transformation rates (1.5 x 10-6 ± 4.0 x 10-7 transformants/total cell number after induction/µg plasmid DNA) as reported previously (Mulder et al., 2017). Integration of linear DNA fragments harboring an antibiotic resistance marker flanked by homologous regions can also be performed by using this protocol. For L. lactis KF147 harboring pNZ6200 natural transformation rates are typically similar to those obtained with pNZ6202 (Mulder et al., 2017).
- GSGM17 (1 L)
M17 medium
2% glucose
0.5 M sucrose (= 170 g)
0.4 M glycine (= 30 g)
Autoclave at 121 °C for 15 min - Washing solution 1 (500 ml)
0.5 M sucrose (= 85 g)
10% v/v glycerol
RO (reversed osmosis) water
Autoclave at 121 °C for 15 min - Washing solution 2 (500 ml)
0.5 M sucrose (= 85 g)
0.05 M EDTA
10% v/v glycerol
RO (reversed osmosis) water
Autoclave at 121 °C for 15 min - Recovery medium
GM17
Note: GM17 is the same as M17 broth but then supplemented with 0.5 % glucose (w/v).
20 mM MgCl2
2 mM CaCl2 - Chemically Defined Medium (CDM) (500 ml)
Note: The recipe for CDM for L. lactis is based on Otto et al., 1983; Poolman and Konings, 1988.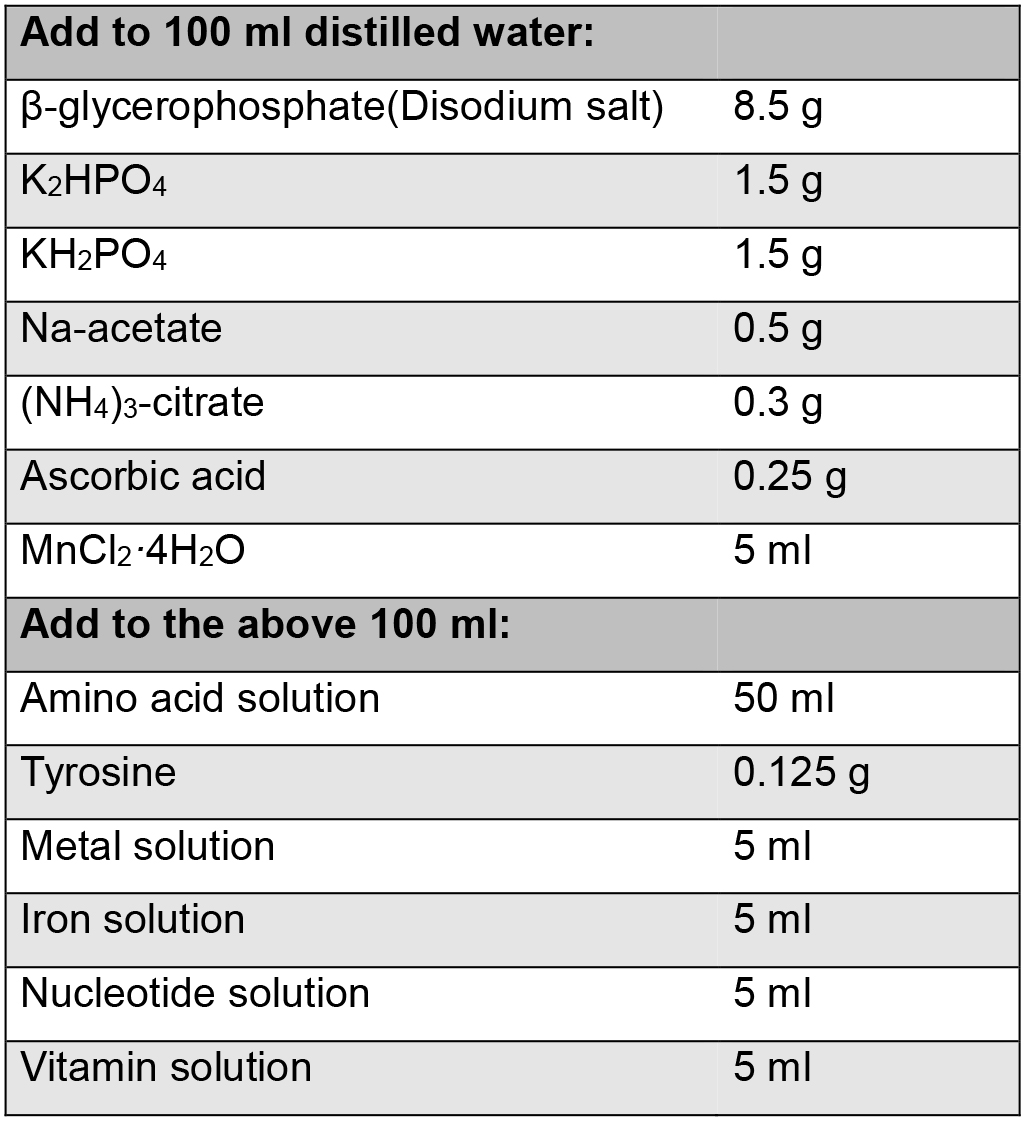
Add up to 500 ml with RO (reversed osmosis) water
Adjust the pH to 6.8
Filter sterilize 500 ml in a sterile 500 ml bottle - GCDM (chemically defined medium supplemented with glucose)
Add sterilized glucose (20% solution, Tritium) to a final concentration of 2% to CDM prior to culturing L. lactis cells to obtain GCDM - Stock solutions for CDM
Note: *Solutions that need to be prepared on the day of use.- Nucleotide solution*
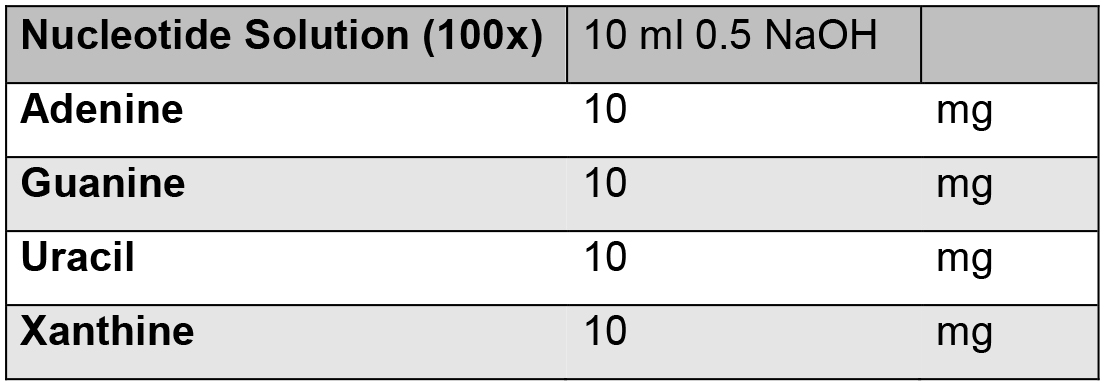
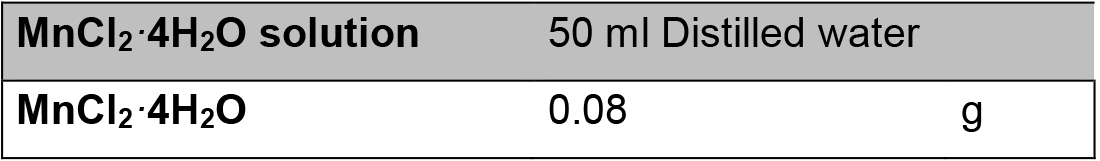
Make fresh and then use 5 ml - MnCl2·4H2O solution*
- Amino acid solution
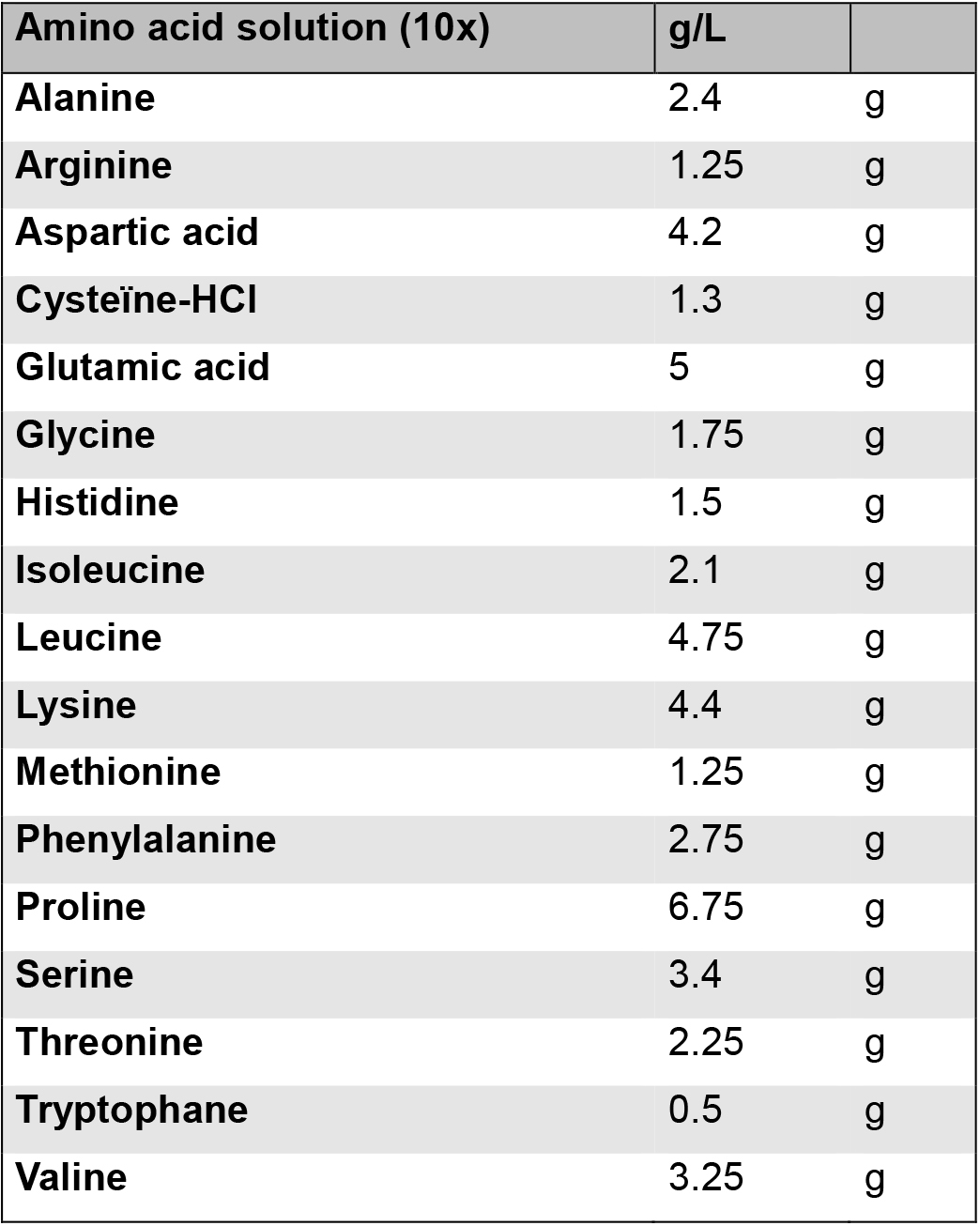
Dissolve at a 6.5 pH (allow 1 h) at room temperature
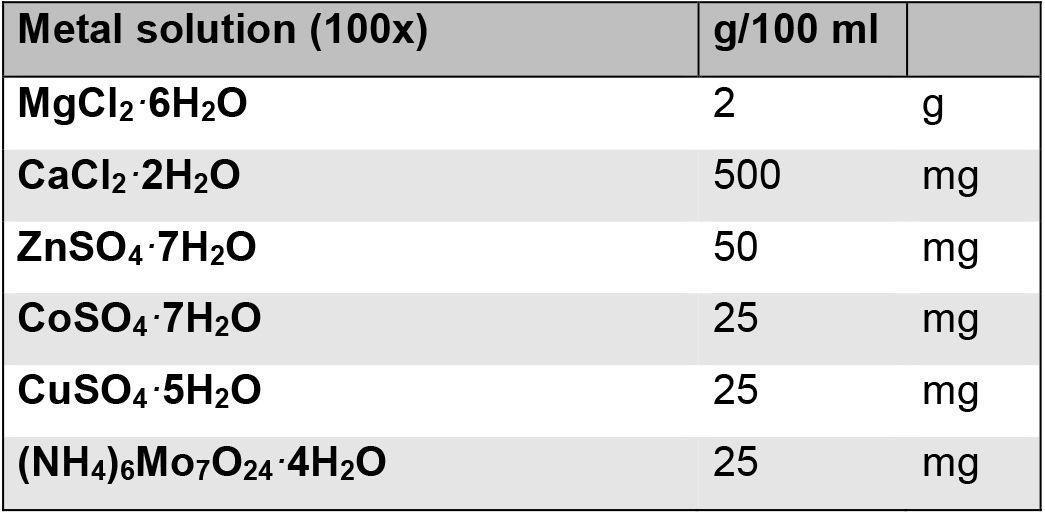
Aliquot into 50 ml CELLSTAR® Polypropylene tubes and store at -20 °C - Metal solution
Aliquot into 6 ml tubes and store at -20 °C - Iron solution

Aliquot into 6 ml tubes and store at -20 °C - Vitamin solution
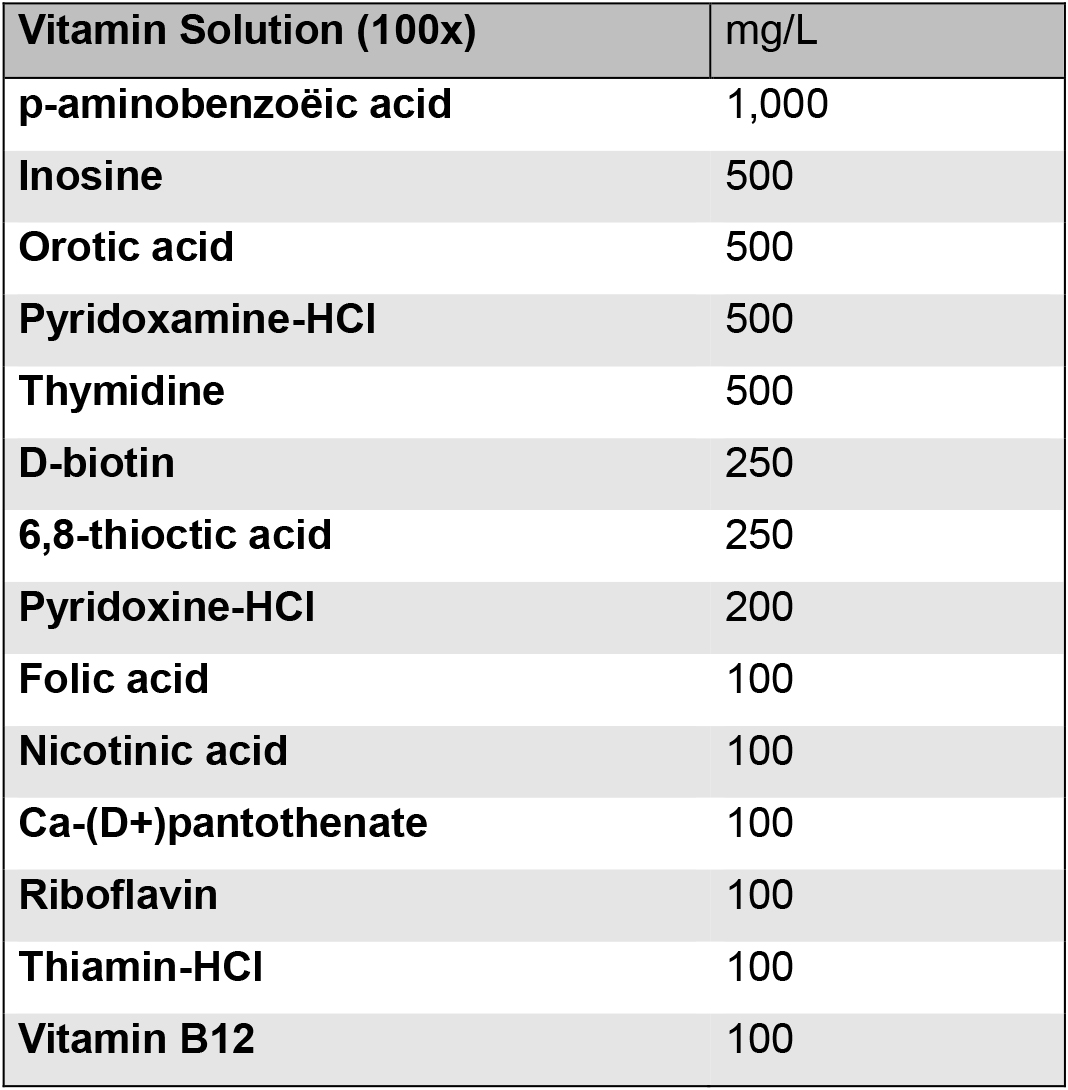
Increase the pH until all components are dissolved. Afterwards, adjust the pH to 7.0
Aliquot into 6 ml tubes and store at -20 °C
- Nucleotide solution*
- Blokesch, M. (2016). Natural competence for transformation. Curr Biol 26(21): R1126-R1130.
- Blomqvist, T., Steinmoen, H. and Havarstein, L. S. (2006). Natural genetic transformation: A novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl Environ Microbiol 72(10): 6751-6756.
- David, B., Radziejwoski, A., Toussaint, F., Fontaine, L., Henry de Frahan, M., Patout, C., van Dillen, S., Boyaval, P., Horvath, P., Fremaux, C. and Hols, P. (2017). Natural DNA transformation is functional in Lactococcus lactis ssp. cremoris KW2. Appl Environ Microbiol.
- Fontaine, L., Dandoy, D., Boutry, C., Delplace, B., de Frahan, M. H., Fremaux, C., Horvath, P., Boyaval, P. and Hols, P. (2010). Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl Environ Microbiol 76(23): 7870-7877.
- Fontaine, L., Wahl, A., Flechard, M., Mignolet, J. and Hols, P. (2015). Regulation of competence for natural transformation in streptococci. Infect Genet Evol 33: 343-360.
- Goel, A., Santos, F., Vos, W. M., Teusink, B. and Molenaar, D. (2012). Standardized assay medium to measure Lactococcus lactis enzyme activities while mimicking intracellular conditions. Appl Environ Microbiol 78(1): 134-143.
- Håvarstein, L. S., Coomaraswamy, G. and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92(24): 11140-11144.
- Horton, R. M., Cai, Z. L., Ho, S. N. and Pease, L. R. (1990). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8(5): 528-535.
- Johnston, C., Martin, B., Fichant, G., Polard, P. and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12(3): 181-196.
- Kleerebezem, M., Beerthuyzen, M. M., Vaughan, E. E., de Vos, W. M. and Kuipers, O. P. (1997a). Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol 63(11): 4581-4584.
- Kleerebezem, M., Quadri, L. E., Kuipers, O. P. and de Vos, W. M. (1997b). Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol 24(5): 895-904.
- Mierau, I. and Kleerebezem, M. (2005). 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol 68(6): 705-717.
- Mulder, J., Wels, M., Kuipers, O. P., Kleerebezem, M. and Bron, P. A. (2017). Unleashing natural competence in Lactococcus lactis by induction of the competence regulator ComX. Appl Environ Microbiol.
- Otto, R., ten Brink, B., Veldkamp, H. and Konings, W. N. (1983). The relation between growth rate and electrochemical proton gradient of Streptococcus cremoris. FEMS Microbiol Lett 16(1): 69-74.
- Pestova, E. V., Havarstein, L. S. and Morrison, D. A. (1996). Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21(4): 853-862.
- Poolman, B. and Konings, W. N. (1988). Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol 170(2): 700-707.
- Seitz, P. and Blokesch, M. (2013). Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37(3): 336-363.
- Wells, J. M., Wilson, P. W. and Le Page, R. W. (1993). Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol 74(6): 629-636.
Data analysis
Notes
Recipes
Acknowledgments
We acknowledge Sabri Cebeci and Koen Giesbers of NIZO for technical assistance. This work was carried out within the BE-Basic R&D Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs. This protocol was adapted from Mulder et al. (2017).The authors have no conflicts of interest to declare.
References
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mulder, J., Wels, M., Kuipers, O. P., Kleerebezem, M. and Bron, P. A. (2018). Induction of Natural Competence in Genetically-modified Lactococcus lactis. Bio-protocol 8(13): e2922. DOI: 10.21769/BioProtoc.2922.
Category
Microbiology > Microbial genetics > Transformation
Molecular Biology > DNA > Transformation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









