- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Brain Tissue Culture of Per2::Luciferase Transgenic Mice for ex vivo Bioluminescence
Published: Vol 8, Iss 13, Jul 5, 2018 DOI: 10.21769/BioProtoc.2917 Views: 7261
Reviewed by: Oneil G. BhalalaKarthik KrishnamurthyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Dual Lentiviral CRISPR-based Transcriptional Activation System for Gene Expression Regulation in Neurons
Katherine E. Savell [...] Jeremy J. Day
Sep 5, 2019 7306 Views

Method for Primary Epithelial Cell Culture from the Rat Choroid Plexus
Valeria Lallai [...] Christie D. Fowler
Feb 20, 2020 6189 Views

Confocal Imaging and 3D Reconstruction to Determine How Genetic Perturbations Impact Presynaptic Morphology at the Mouse Calyx of Held
Christian Keine [...] Samuel M. Young Jr.
Sep 5, 2023 1880 Views
Abstract
In circadian research, it is essential to be able to track a biological rhythm for several days with the minimum perturbation for the organisms or tissues. The use of transgenic mice lines, in which the luciferase reporter is coupled to a molecular clock protein (here PERIOD2), gives us the opportunity to follow the circadian activity in different tissues or even single clock cells for days without manipulation. This method creates sections using a mouse brain matrix, which allows us to obtain several brain samples quickly at a single time point.
Keywords: BioluminescenceBackground
Circadian rhythms are behavioral or molecular changes that follow roughly 24 h-cycles and are sustained without any external cue. In mammals, locomotor activity, body temperature and hormone release are examples of circadian rhythms which are under the control of the suprachiasmatic nucleus (SCN) clock located in the hypothalamus. The ability of the SCN cells to keep an endogenous rhythm is due to a molecular machinery composed by the positive and negative loops of the expression of clock genes: firstly, CLOCK and BMAL1 proteins heterodimerize to activate the transcription of different genes through E-box sites on the promoter which is on genes like period (Per1-3) and cryptochrome (Cry1-2; Takahashi et al., 2008). Then, the proteins of PERIOD and CRYPTOCHROME heterodimerize and enter back to the nucleus to prevent BMAL1 binding to the E-Box. Hence, PERIOD and CRYPTOCHROME inhibit their own transcription (Takahashi et al., 2008). A second loop is made by retinoid-related orphan receptors (ROR) and Rev-Erb: the ROR proteins activate Bmal1 gene while REV-ERB proteins inhibit it via ROR-response element in the Bmal1 promoter. All this mechanism oscillates within a 24 h-period (Takahashi et al., 2008).
In circadian research, it is important to follow rhythmic activity in the whole organism or tissues around the 24 h. For that, it is necessary to get tissues or samples at different time points to model the oscillations of gene, protein expression or hormonal release. However, these methods require more than one animal per time point, and therefore it requires a lot of animals to get a complete and significant circadian oscillation.
In 2000, Yamazaki et al., created a transgenic rat line to solve this problem. They inserted a vector containing the luciferase gene from the firefly under the control of Per1 promoter. Since the 80’s, the luciferase has been used as ATP, gene or protein reporter. This 61-kDa enzyme has the particularity to release photon by oxidation of its substrate and in the presence of ATP, Mg2+ and oxygen (Gould and Subramani, 1988). The beetle luciferase has the advantage to be a single protein with no post-translational modification; its catalytic area is ready-to-use after its translation and minimal auto-fluorescence throughout recording (Bioluminescent Reporters [Reference #1]).
Although Per1-luciferase rat is an advance in circadian field, it does not allow us to follow the endogenous clock gene expression, but rather the endogenous activity of the heterodimer CLOCK-BMAL1. In 2004, Takahashi lab created the transgenic mouse line in which the open reading frame (ORF) of the luciferase is fused to the end of the Per2 gene (Yoo et al., 2004). All cells expressing the PER2 protein are also able to produce yellow-green light (~560 nm) in the absence of external light source if they have access to the luciferin: the consumable substrate. The bioluminescence produced by these cells permits to follow the circadian clock activity of the same individual for several days, and even weeks.
The principal aim of this technique is to dissect the brain region of interest of several animals at a single time point. For that, we used a mouse brain matrix that requires less tissue preparation and slices faster than a vibratome. However, the disadvantage of this technique is the loss of thickness precision (~500 µm). The vibratome cuts thinner and more precise tissue slices, but all the related procedure requires time. The advantage of the use of the matrix is to have few steps to work rapidly on the area of interest.
Materials and Reagents
- Gloves
- Carbon steel scalpel No. 24 (Swann-Morton, catalog number: 0211 )
- Double edge stainless steel razor blade (Electron Microscopy Sciences, catalog number: 72000 )
- NuncTM cell culture/Petri dishes (35 mm, Thermo Fisher Scientific, catalog number: 150318 )
- 10 ml syringe (Terumo Medical, catalog number: SS-10ES )
- Corning® vacuum filter system 500 ml, sterile, pore size: 0.22 µm (Corning, catalog number: 431097 )
- Sterile sampling pot: aseptic 40 ml polypropylene straight container with screw cap (Dominique DUTSCHER, Corning GOSSELINTM, catalog number: 688252 )
- Falcon Corning® 15 ml PP Centrifuge Tubes (Corning, catalog number: 430791 )
- Falcon Corning® 50 ml PP Centrifuge Tubes (Corning, catalog number: 430829 )
- FisherbrandTM SureOneTM 0.1-10 µl aerosol barrier pipette tips (Fisher Scientific, catalog number: 11903466 )
- FisherbrandTM SureOneTM 20-200 µl aerosol barrier pipette tips (Fisher Scientific, catalog number: 11963466 )
- FisherbrandTM SureOneTM 100-1,000 µl aerosol barrier pipette tips (Fisher Scientific, catalog number: 11973466 )
- Costar® 5 ml Stripette® serological pipets, sterile (Corning, catalog number: 4487 )
- Costar® 10 ml Stripette® serological pipets, sterile (Corning, catalog number: 4488 )
- Costar® 25 ml Stripette® serological pipets, sterile (Corning, catalog number: 4489 )
- Axygen® 0.2 ml Thin Wall PCR Tubes with Flat Cap (Corning, catalog number: PCR-02-C )
- Axygen® 0.6 ml Maxy Clear SnaplockMicrocentrifuge Tube (Corning, catalog number: MCT-060-C )
- Millicell® cell culture insert (Merck, catalog number: PICMORG50 )
- Per2::Luciferase homozygote knock-in (KI) Musmusculus (Per2tm1Jt)
Note: Mice were initially from Jackson Laboratories. Generally, we used young-adult (2-6 months old) mice, males as well as females, from our own breeding colony (Chronobiotron platform, UMS-3415 in Strasbourg). Aside from specific protocols, mice were housed in groups–of 3 or 4 individuals with food and water available ad libitum– in light-proof ventilated rooms, under 12 h white light and 12 h dim red light (< 5 lux at cage level) cycle (LD12:12; lights on at 7:00 A.M.). - Antibiotic (penicillin-streptomycin 10,000 U/ml; 10,000 mg/ml, Sigma-Aldrich, catalog number: P4333 ), stored at -20 °C
- B27 (Thermo Fisher Scientific, catalog number: 17504044 ), stored at -20 °C
- Hank’s balanced salt solution with red phenol (HBSS; 10x, Sigma-Aldrich, catalog number: H1641 ) stored at room temperature
- HEPES (Sigma-Aldrich, catalog number: H0887 ), stored at 4 °C
- Sodium bicarbonate (Sigma-Aldrich, catalog number: S8761 ), stored at 4 °C
- Bactericide/fungicide: ANIOSYME DD1 (Laboratoires ANIOS, Lille-Hellemmes, France)
- Commercial chlorine in tablets to make 10x bleach
- Dulbecco's modified Eagle's medium 10x (DMEM) with low glucose without red phenol (Sigma-Aldrich, catalog number: D2902 ), stored at 4 °C
- D (+) Glucose (Sigma-Aldrich, catalog number: G7021 ), stored at room temperature
- Beetle luciferin (Promega, catalog number: E1602 ), stored at -80 °C
- High vacuum grease (Dow corning®, Wiesbaden, Germany), stored at room temperature
- MilliQ Water
- EtOH 70%
- 0.1 M luciferin (see Recipes), stored at -20 °C
- HBSS 1x (see Recipes), stored at 4 °C
- DMEM 1x (see Recipes), stored at 4 °C
- Cleaning solution for tools (see Recipes)
Equipment
Note: All items without reference can be ordered from any qualified company.
- Stainless steel mouse brain matrix (Adult Mouse Brain Slicer Matrix, Zivic Instruments, catalog number: BSMAS005-1 )
Note: Other brain matrixes (for hamsters or rats) exist. - Stainless steel tweezers, fine tips, straight
Note: To have a better sight of the dissection tools, see Figure 1.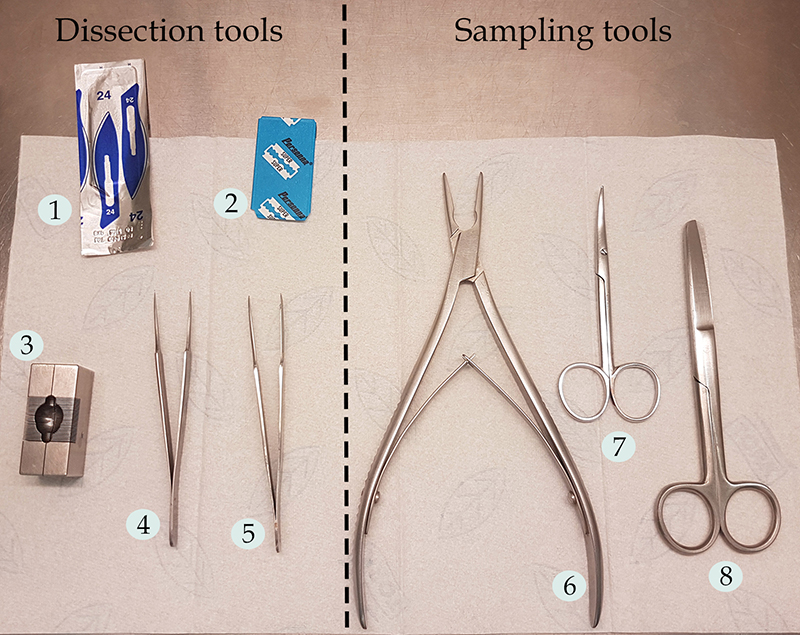
Figure 1. Surgery instruments used during the protocol. 1) Carbon steel scalpel No. 24; 2) Double edge stainless steel razor blade; 3) Stainless steel mouse brain matrix; 4-5) Stainless steel tweezers, fine tips, straight; 6) Friedman bone rongeur; 7) Curved Scissors, fine tips; 8) Stainless steel operating scissors, straight. - Stainless steel operating scissors, straight
- Curved Scissors, fine tips
- Friedman bone rongeur
- Vacuum pump (KNF, catalog number: N86KN.18 )
- Red light lamp (around 620-650 nm)
- Fiber optic light source (SCHOTT, catalog number: KL 1500LCD )
- Stereo Microscope (Nikon, catalog number: 536087 )
- Sterilized BRAND® Petri dish, glass, size 100 mm x 15 mm (BRAND, catalog number: 455742 )
- 1 L sterilized bottle
- 500 ml sterilized bottle
- 100 ml sterilized measuring cylinder
- 1 L sterilized measuring cylinder
- 1 L sterilized beaker
- Magnet
- Magnetic plate (size: big enough for 1 L beaker; strength: enough to dissolve powder into a liquid)
- P10 pipetman® classic (Gilson, catalog number: F144802 )
- P20 pipetman® classic (Gilson, catalog number: F123600 )
- P200 pipetman® classic (Gilson, catalog number: F123601 )
- P1000 pipetman® classic (Gilson, catalog number: F123602 )
- Pipetboy acu 2 (INTEGRA Biosciences, catalog number: 155019 )
- Milli-Q® water system
- Ice maker
- Fridge (4 °C)
- Fume hood (horizontal flow)
- Fume hood (vertical flow)
- Autoclave
- Photo-counting apparatus as LumiCycle 32 (Actimetrics, Wilmette, IL, USA) inside 36.5 °C incubator
- 37 °C incubator
Note: Petri dishes will be sealed. We do not need 5% CO2; an incubator that keeps only medium or dishes at 37 °C is enough.
Software
- LumiCycle Analysis for data extraction
- Table software like Microsoft Office Excel for data extraction
- SigmaPlot for statistics
- GraphPad for graphs
Procedure
Before the experiment
- Prepare antibiotic and B27 aliquots, and keep them at -20 °C. Prepare 1 mM of luciferin (see Recipes).
Note: The aliquots should avoid repeat freeze/thaw cycles, optimally less than 5 times. - Classify your animals according to their genotype to select homozygote knock-in (KI) mice for the transgene Per2::Luc; keep in mind that the heterogeneous KI/+ should have the half of the signal due to the presence of only one allele.
Note: Probes that we used for PCR genotyping are the same as that described in Yoo et al. (2004). - Prepare HBSS 1x and DMEM 1x under a clean fume hood (see Recipes).
Note: Fresh HBSS 1x and DMEM 1x can be prepared on the day of experiment, but HBSS has to be cold enough.
On the day of the experiment
- Preparation
- Prepare the DMEM medium ready-to-use by adding B27, antibiotics and luciferin (see Recipes).
Note: It is not necessary to filter the medium again. - Prepare sampling pot, for each animal, filled with HBSS 1x enough for the brain to bathe (around 10 ml).
- Prepare culture dishes as many as samples. Number them on the cover side. Put 1 ml of ready-to-use DMEM in each and cover the top with vacuum grease. The grease avoids the medium to evaporate at 37 °C: the samples will not dry and the apparatus will stay dry. Put the cover gently on its Petri dish and keep them at 37 °C in the incubator.
Notes:- Avoid red/pink ink. To open the LumiCycle and put the samples inside, it must be done under red light.
- To easily cover the top, first fill a 10 ml syringe with the grease (see Video 1).
- Avoid red/pink ink. To open the LumiCycle and put the samples inside, it must be done under red light.
Video 1. Petri dish sealing. This video shows how we use a syringe to seal Petri dishes cleanly.- Clean the fume hood (horizontal flow) and all the equipment inside (stereomicroscope, lamp) with 70% EtOH. Clean the brain matrix and all the forceps with 70% EtOH and let them dry under the clean fume hood. Cover the brain matrix with aluminum paper and keep it at 4 °C at least 1 h before to cool it down.
Note: It is also possible to put the matrix in the freezer to cool it faster but do not let it inside too much time. If the matrix is too cold, the brain will stick on it. If it happens, put some drops of HBSS medium on the brain and the matrix to take off the slices.
- Prepare the DMEM medium ready-to-use by adding B27, antibiotics and luciferin (see Recipes).
- Sampling
- Kill the mouse by cervical dislocation.
- Then, obtain the brain quickly and keep it in HBSS 1x on ice/or in the fridge until dissection.
- To avoid degradation of the tissue (if all the procedure reaches more than ~1 h), we recommend starting with the sacrifice of one or two animals at once.
Notes:- This sacrifice method is performed in accordance with the rules of the French National Law and the European Committee Council Directive of September 22, 2010 (2010/63/UE).
- We do not use sedation. You can use another method of sacrifice that allows you to obtain quickly the brain.
- If you need more than one sample, be careful at what time you are sampling. It is important for sampling at the same moment, or to check if sampling at different moment of the day is not affecting your data.
- We generally spent less than 1 min per animal to extract the brain after its death.
- This sacrifice method is performed in accordance with the rules of the French National Law and the European Committee Council Directive of September 22, 2010 (2010/63/UE).
- Kill the mouse by cervical dislocation.
- Dissection
- Put the brain inside the matrix and slice it in a single act with the razor blade (around 500 µm of thickness). To place the brain upside down could be a solution to find anatomical landmarks as the optic chiasma, and find the location of the slice containing the area of interest with the help of a brain atlas (see Figure 2). It is also possible to section the whole brain (see Video 2) and, afterward select the slice where your structure of interest is.
Note: We generally spent less than 2 min to slice the whole brain.
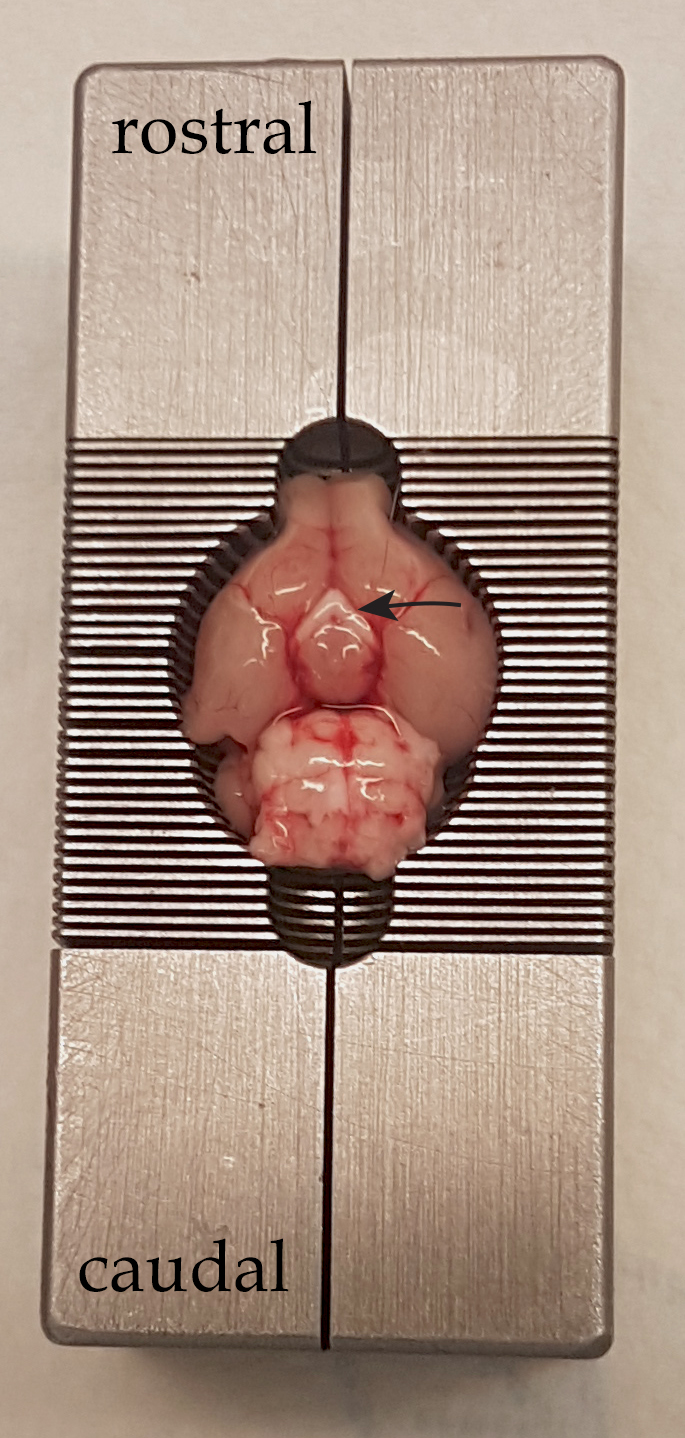
Figure 2. Upside down mouse brain in the matrix. The optic chiasma pointed with the arrow can be used as an anatomical landmark.Video 2. Brain slicing with the mouse matrix- Microdissection is done under the stereomicroscope in cold HBSS, in the glass and sterilized Petri dish. Try to minimize the sample to fit in 1 mm2: in this way, tissue will have access to all the nutrients needed easily, and avoid necrosis.
Notes:- It is also possible to use plastic culture dish, but after one or two microdissections, the scalpel leaves marks on it while it does not on glass. Also, glass Petri dish can be used again after sterilization.
- We usually use forceps to maintain the sample and scalpel to cut a square containing the structure of interest. However, it is important to microdissect only the area of interest if adjacent areas also oscillate. For example, the SCN fits in a square without any problem, while another brain structure of our interest, the habenula, needs to have all the other tissues around it removed.
- We generally spent between 3-5 min to microdissect according to the difficulty of the area of interest. If you have many brains, one person can slice the brain with the matrix while the second one is microdissecting to accelerate the experiment.
- It is also possible to use plastic culture dish, but after one or two microdissections, the scalpel leaves marks on it while it does not on glass. Also, glass Petri dish can be used again after sterilization.
- Pick up the culture dish inside the incubator corresponding to the sample and put the cell culture insert (Millicell®) on the medium. It should not have any bubbles under it. If this is the case, take it out and put it back. Place your sample on the top of it with your forceps or by recovering your sample with your 1,000 µl pipette. Remove the remained HBSS1x on the top of the culture insert (see Video 3).
Video 3. Sample dropping on the Millicell® culture insert
- Return the sample into the incubator until you collect all of them.
Note: We generally spent less than 5 min to place the sample on the insert and to return the petri dish inside the incubator.
- Put the brain inside the matrix and slice it in a single act with the razor blade (around 500 µm of thickness). To place the brain upside down could be a solution to find anatomical landmarks as the optic chiasma, and find the location of the slice containing the area of interest with the help of a brain atlas (see Figure 2). It is also possible to section the whole brain (see Video 2) and, afterward select the slice where your structure of interest is.
- LumiCycle
- At the end, put your samples in a photon-counting luminometer as the LumiCycle. You will need to be in dim red light and in low intensity to avoid the light (mostly green-yellow light) altering sensitive photomultipliers inside. The LumiCycle 32, as its name says, allows you to have 32 samples inside. You can turn on them at once or at several times.
- We usually record our sample 3 times in one hour by setting 112 sec of recording per sample.
- After 7 days, you will need to change your medium if you want to keep the sample on recording.
Note: It depends on your sample (the size, its consumption of luciferin…) and your experiment schedule. You need to check your sample oscillations and change them when they are low.
- At the end, put your samples in a photon-counting luminometer as the LumiCycle. You will need to be in dim red light and in low intensity to avoid the light (mostly green-yellow light) altering sensitive photomultipliers inside. The LumiCycle 32, as its name says, allows you to have 32 samples inside. You can turn on them at once or at several times.
- Cleaning
- To make sure that no bacteria or fungi will grow in medium used for dissection or culture, we add 10x bleach into it until the liquid becomes clear before throwing away.
- Place all the forceps used and the brain matrix in the cleaning solution for 20 min minimum. They need to be fully dipped or at least the part that was in contact with the tissues. Wash the equipment under tap water and dry them. Be careful the solution is toxic; therefore, do not discard it in the sink.
Note: We usually let the tools in the cleaning solution overnight. Avoid bleach as it will harm your equipment. - Clean the fume hood (horizontal flow) and all the equipment inside (stereo microscope, lamp) with 70% EtOH.
- To make sure that no bacteria or fungi will grow in medium used for dissection or culture, we add 10x bleach into it until the liquid becomes clear before throwing away.
Data analysis
- LumiCycle Analysis
Note: You will be able to copy the recording file(s) at every moment of the experiment even if the LumiCycle is still recording.
We use the LumiCycle analysis which fits a mathematical model for the extraction of the oscillation characteristics (period, amplitude, acrophase). You can calculate manually, but the software is quite accurate with less effort.
First, we subtract the baseline of our oscillations with 24 h-running average. In the end, we obtain a sinusoid curve which oscillates around the 0 of the y-axis which can be associated to different mathematical models. The smoothing of the data helps for a best fitting between the experimental and theoretical oscillations.
Then, we use the ‘LM fit (dampened)’ model which fits best to our oscillation. It takes the dampening into account and gives us a proper fit (goodness), meaning that 100% is a perfect fitting of your model on your data (LumiCycle software, Actimetrics). From the mathematical model, the software calculates the period and the first peak amplitude (by selecting the relative T0) of the selected time window of the oscillation. Unlike the ‘LM fit’, periodogram model calculates the mean of the period and amplitude of the oscillation along time. We also measure the acrophase, the time when the peak occurs, with the help of the yellow slider which gives us the time at its position.
Refer to Video 4 for the detailed procedures.
Note: We usually skip the first hours of recording due to the huge peak of bioluminescence at the beginning (artifact). We recommend doing this analysis on at least 3-days oscillations.Video 4. Data analysis step by step with the LumiCycle Analysis software. This video shows how to use the data file given by the LumiCycle, how to analyze the data with the software tools and how to extract the data on Excel software to have a look into the oscillation.
Then, we export oscillation data (raw or baseline subtracted) as .csv file which can be opened by a table software. This allows us to recreate the oscillation using the graph software desired (see Figure 3).
Figure 3. Example of an oscillation of habenula sample from a Per2::Luciferase transgenic mouse. After 3 days (first arrow), the sample was removed from the LumiCycle for another experiment and it was put back with a new and fresh culture medium (second arrow). The last arrow indicates that the sample is dying after the last medium change (around 10.5 days). - Exclusion
The ‘LM fit (dampened)’ model offers a proper fit (goodness). A threshold should be defined by the user under which the oscillation is considered to be arrhythmic. However, the sample cannot be excluded.
When no rhythm is observed and the data is like flat, there are two possibilities: cells are desynchronized (at the tissue level, the rhythm disappears) or the sample is dead. When a sample is dead, the data values of bioluminescence have to be in the range of the noise of the apparatus. The noise can be obtained by putting just the medium without any sample in the LumiCycle for each photomultiplicator.
Note: We usually placed the threshold at 70% for habenula samples. You need to verify each time how your data fits the model.
Notes
Troubleshooting steps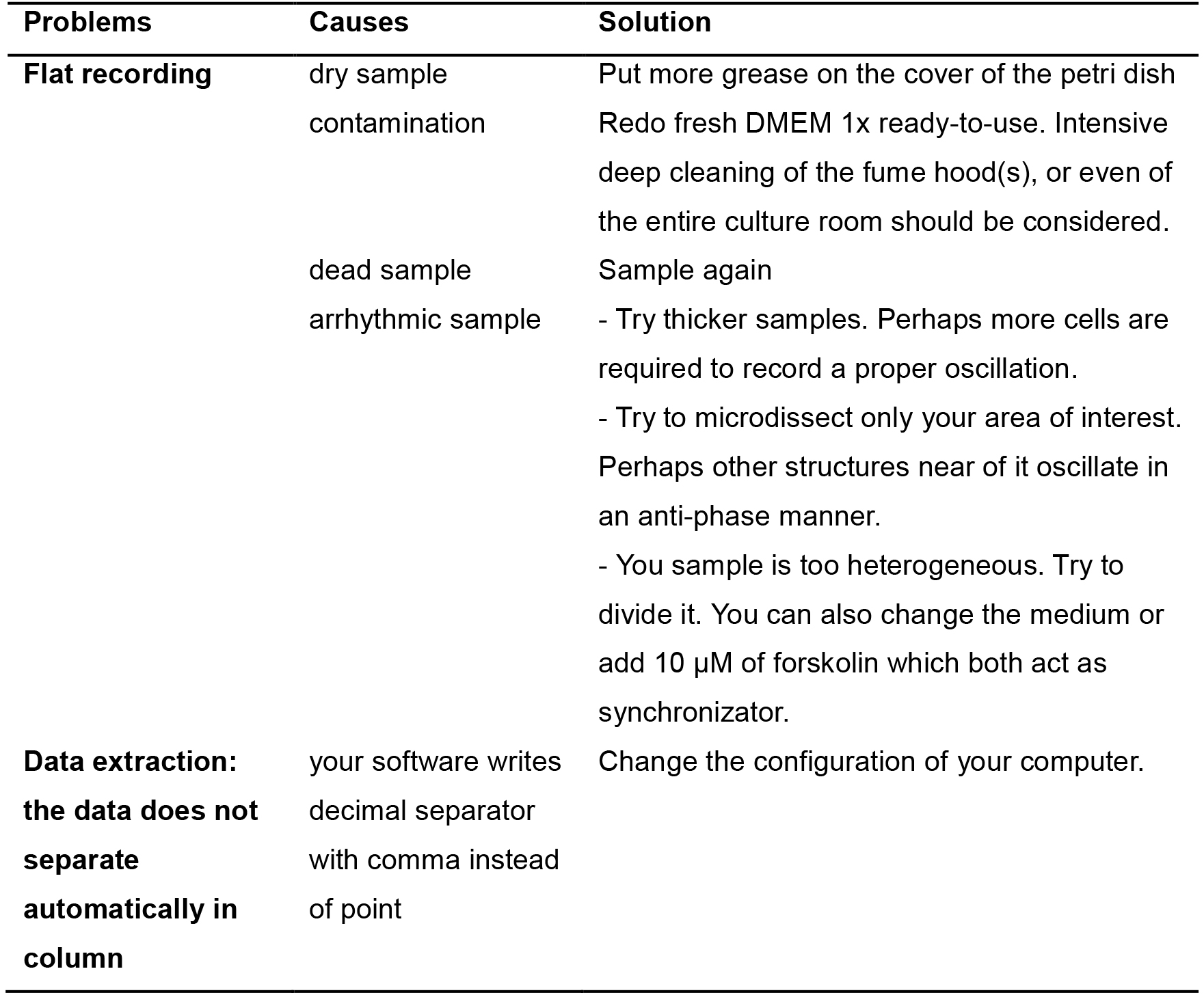
Recipes
- 0.1 M luciferin
Note: In this protocol, we used 0.1 mM luciferin. Too high concentration is toxic. Other protocols can be used for different concentrations of luciferin but they should not exceed 1 mM. Be aware that different concentrations of luciferin will lead to different results. For detailed information, refer to Feeney et al. (2016).- Add 1.57 ml of MilliQ water directly in the luciferin jar (50 mg)
- Prepare aliquots of it and store them at -20 °C in light proof container
- Add 1.57 ml of MilliQ water directly in the luciferin jar (50 mg)
- HBSS 1x (for 1 L)
- Add all the component in a sterilized beaker under a clean fume hood (use sterilized materials/equipment only):
100 ml of HBSS 10x
4.7 ml of sodium bicarbonate
10 ml of HEPES
10 ml of antibiotics
Complete with sterile MilliQ water - Keep it cold in 1 L sterilized bottle in the fridge. You can use it up to 4-5 months if it is still clear/clean
- Add all the component in a sterilized beaker under a clean fume hood (use sterilized materials/equipment only):
- DMEM 1x (for 1 L)
- Add all the components in a sterilized beaker under a clean fume hood (use sterilized materials/equipment only):
10 g of DMEM 10x (all the powder of the jar)
4.7 ml of sodium bicarbonate
10 ml of HEPES
1.5 ml of antibiotics
3.5 g of D (+) glucose
500 ml of sterile MilliQ water - Homogenize until the powder is fully dissolved by using a clean magnet and complete it with sterile MilliQ water
- Filtrate (0.22 μm, sterile) and store it in at least 2 sterilized 500 ml bottles. We use them up in 4 months since the bottle is opened if the medium remains clear/clean; 2 bottles will be available for 8 months
- On the experiment day, for each sample, prepare 1 ml of DMEM 1x and add 20 µl of B27, 1 µl of antibiotics and 1 µl of luciferin (stock solution 0.1 M) just before use. Keep it at 37 °C in a light proof place
Note: We usually prepare 1 ml extra. For 7 samples, prepare 8 ml in a Falcon tube.
- Add all the components in a sterilized beaker under a clean fume hood (use sterilized materials/equipment only):
- Cleaning solution
25 ml of ANIOSYME DD1 in 5 L of tap water
Acknowledgments
The authors have no conflict of interest to declare. Funding sources of the present study were provided by the Agence National de la Recherche (ANR-14-CE13-0002-01 ADDiCLOCK JCJC to JM and NLS Ph.D. fellow) and the Centre National de la Recherche Scientifique (JM).
The authors thank Mr. Guillaume Vanotti for his help with the video production, Dr. Nadia Mazzaro and Mr. Maxime Sartori for their comments and suggestions and Mr. Hikmet Undemir for the proofreading.
This protocol was adapted from the procedures published in Yoo et al. (2004) and Yamazaki and Takahashi (2005).
References
- Bioluminescent Reporters. Promega. (Accessed January 9, 2018)
- Feeney, K. A., Putker, M., Brancaccio, M. and O'Neill, J. S. (2016). In-depth characterization of firefly luciferase as a reporter of circadian gene expression in mammalian cells. J Biol Rhythms 31(6): 540-550.
- Gould, S. J., and Subramani, S. (1988). Firefly luciferase as a tool in molecular and cell biology. Anal Biochem 175: 5-13.
- Takahashi, J. S., Hong, H. K., Ko, C. H. and McDearmon, E. L. (2008). The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9(10): 764-775.
- Yamazaki, S. and Takahashi, J. S. (2005). Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol 393: 288-301.
- Yamazaki, S., Numano, R., Abe, M., Hida, A., Takahashi, R., Ueda, M., Block, G. D., Sakaki, Y., Menaker, M. and Tei, H. (2000). Resetting central and peripheral circadian oscillators in transgenic rats. Science 288(5466): 682-685.
- Yoo, S. H., Yamazaki, S., Lowrey, P. L., Shimomura, K., Ko, C. H., Buhr, E. D., Siepka, S. M., Hong, H. K., Oh, W. J., Yoo, O. J., Menaker, M. and Takahashi, J. S. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101(15): 5339-5346.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Salaberry, N. L. and Mendoza, J. (2018). Brain Tissue Culture of Per2::Luciferase Transgenic Mice for ex vivo Bioluminescence. Bio-protocol 8(13): e2917. DOI: 10.21769/BioProtoc.2917.
Category
Neuroscience > Cellular mechanisms > Tissue isolation and culture
Cell Biology > Tissue analysis > Tissue recording
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link