- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Intact Vacuoles from Petunia Petals and Extraction of Sequestered Glycosylated Phenylpropanoid Compounds
(*contributed equally to this work) Published: Vol 8, Iss 13, Jul 5, 2018 DOI: 10.21769/BioProtoc.2912 Views: 8162
Reviewed by: Amey RedkarBen SpitzerGongjun Shi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Extraction and Measurement of Strigolactones in Sorghum Roots
Kaori Yoneyama [...] Koichi Yoneyama
Mar 20, 2016 11210 Views

Metabolite Profiling of Mature Arabidopsis thaliana Seeds Using Gas Chromatography-Mass Spectrometry (GC-MS)
Hagai Cohen [...] Rachel Amir
Nov 5, 2016 12494 Views

Analysis of Modified Plant Metabolites Using Widely Targeted Metabolite Modificomics
Jianing Zhang [...] Jun Yang
Apr 5, 2025 1458 Views
Abstract
Plant vacuoles are the largest compartment in plant cells, occupying more than 80% of the cell volume. A variety of proteins, sugars, pigments and other metabolites are stored in these organelles (Paris et al., 1996; Olbrich et al., 2007). Flowers produce a variety of specialized metabolites, some of which are unique to this organ, such as components of pollination syndromes, i.e., scent volatiles and flavonoids (Hoballah et al., 2007; Cna'ani et al., 2015). To study the compounds stored in floral vacuoles, this compartment must be separated from the rest of the cell. To enable isolation of vacuoles, protoplasts were first generated by incubating pierced corollas with cellulase and macrozyme enzymes. After filtering and several centrifugation steps, protoplasts were separated from the debris and damaged/burst protoplasts, as revealed by microscopic observation. Concentrated protoplasts were lysed, and vacuoles were extracted by Ficoll-gradient centrifugation. Vacuoles were used for quantitative GC-MS analyses of sequestered metabolites. This method allowed us to identify vacuoles as the subcellular accumulation site of glycosylated volatile phenylpropanoids and to hypothesize that conjugated scent compounds are sequestered in the vacuoles en route to the headspace (Cna'ani et al., 2017).
Keywords: VacuoleBackground
Plant vacuoles occupy up to 80% of the cellular volume in plant cells. These organelles are essential for plant growth and development, with varied functions throughout the plant's life. Vacuoles compartmentalize different components, such as proteins, sugars, ions and specialized metabolites, and are involved in the plant's response to different developmental and environmental signals, e.g., stomatal opening, adaptation to cold, defense against herbivores and floral pigmentation (Shitan and Yazaki, 2013). Specific transporters are employed by vacuoles to allow penetration of inorganic ions and hydrophilic metabolites through the lipid bilayer membrane of the tonoplast (Schneider et al., 2012; Liu et al., 2015).
Specialized metabolites, including floral scent volatiles, are often produced and/or accumulated in sink organs, such as flower petals, glandular trichomes, root bark, etc. (Hanhineva et al., 2008; Kortbeek et al., 2016; Lashbrooke et al., 2016). Volatile phenylpropanoids and other specialized metabolites, e.g., flavonoids, monoterpenes, betalains, alkaloids and brassinosteroids, undergo various postproduction modifications, such as glycosylation, methylation and acylation. These modifications increase their stability, enable transport, lower their toxicity by blocking reactive groups and enhance their water solubility, thus enabling storage in subcellular compartments (Bowles et al., 2005; Dean et al., 2005). Glycosylated scent compounds are generally regarded as storage forms or precursors for the emission of aglycones at the appropriate time or stage of plant or organ development (Rambla et al., 2014). Floral scent has been studied extensively in the model plant Petunia. However, little is known about the intracellular fate of scent compounds. To this end, based on several previously described protocols from different plants/tissues (Robert et al., 2007; Fontes et al., 2010; Faraco et al., 2011 and 2014; Pérez-Díaz et al., 2014; Shen et al., 2014; Cna'ani et al., 2017), we generated a procedure for vacuolar isolation from petunia petals. Using a GC-MS–based protocol for isolating glycosylated volatiles from petal vacuoles, we were able to reveal a mechanism used by flowers to sequester volatile phenylpropanoids in vacuoles prior to their developmentally regulated emission to the environment.
Materials and Reagents
- Aluminum foil
- Cell strainer 100 μm nylon (Corning, catalog number: 431752 )
- Centrifuge tube 15 ml (Corning, catalog number: 430790 )
- Centrifuge tube 50 ml (Corning, catalog number: 430828 )
- Corning® bottle-top vacuum filter system (Corning, catalog number: 430796 )
- Cover slides 24 x 50 mm (Knittel Glass, catalog number: VD1 2450 Y100A )
- Cap for 4ml vial (J.G. Finneran, catalog number: 5360-13 )
- Finnpipette® pipette tips 5 ml (Sigma-Aldrich, Thermo Fisher, catalog number: P2924 )
- Glass insert (J.G Finneran, catalog number: 401BS-530 )
- Microscope slides 3 x 1 inch (Knittel Glass, catalog number: VA31100 001FKB )
- Non-sterile scalpel blades (Bar Naor, catalog number: BN10011-00 )
- Paper sheets (Romical, catalog number: 322-05004040 )
- PARAFILM® M sealing film (BRAND, catalog number: 701605 )
- Pasteur pipettes open tip 150 mm (Hilgenberg, catalog number: 3150101 )
- Petri dishes 90 x 15 mm (MINIPLAST, catalog number: 820-090-01-017 )
- Safe-Lock Tubes, 1.5 ml, Eppendorf QualityTM, colorless (Eppendorf, catalog number: 0030120086 )
- Safe-Lock Tubes, 2.0 ml, Eppendorf QualityTM, colorless (Eppendorf, catalog number: 0030120094 )
- Cap for 2 ml Vial (CHROMSERVIS, catalog number: 1076-8002-C )
- Silicone tubing 8 x 12 mm (Chen Samuel, catalog number: 054080120 )
- Sterile pipette 1 ml (Alexred, catalog number: ALP ON1E1 )
- Vial 2 ml (J.G Finneran, catalog number: 32008-1232 )
- Vial 4 ml (J.G. Finneran, catalog number: 34013-1545 )
- Ammonium nitrate (Sigma-Aldrich, catalog number: 256064 )
- Calcium chloride dihydrate (Merck, catalog number: 102382 )
- Cellulase R10 (Duchefa Biochemie, catalog number: C8001 )
- D-Mannitol (Sigma-Aldrich, catalog number: M4125 )
- Ethylenediaminetetraacetic acid disodium salt dihydrate (Sigma-Aldrich, catalog number: E6635 )
- Ficoll® PM 400 (Sigma-Aldrich, catalog number: F4375 )
- Fluorescein diacetate solution (Sigma-Aldrich, catalog number: F7378 )
- Gamborg’s B-5 Basal Salt Mixture (Sigma-Aldrich, catalog number: G5768 )
- HEPES (Sigma-Aldrich, catalog number: H7006 )
- Hydrochloric acid 37% (w/w) (Bio Basic, catalog number: HC6025 )
- Isobutyl benzene (Sigma-Aldrich, catalog number: 113166 )
- KCl (Sigma-Aldrich, catalog number: P4504 )
- Macrozyme R10 (Duchefa Biochemie, catalog number: M8002 )
- MES sodium salt (Sigma-Aldrich, catalog number: M3885 )
- Methyl alcohol (Sigma-Aldrich, catalog number: 322415 )
- n-Hexane HPLC (Biosolve, catalog number: 082906 )
- Sodium citrate tribasic dihydrate (Sigma-Aldrich, catalog number: C8532 )
- Sodium hydroxide (Sigma-Aldrich, catalog number: 221465 )
- Sodium hypochlorite 5% (Romical, catalog number: 73-7586-1400 )
- Sodium phosphate dibasic (Sigma-Aldrich, catalog number: 255793
- Sodium phosphate dibasic heptahydrate (Sigma-Aldrich, catalog number: S2429 )
- Sodium phosphate monobasic monohydrate (Sigma-Aldrich, catalog number: S3522 )
- Sterile double-distilled water (DDW)
- Sucrose (DAEJUNG CHMICAL & METALS, catalog number: 7501-4400 )
- Viscozyme (cellulolytic enzyme mixture) (Sigma-Aldrich, catalog number: V2010 )
- TEX buffer (see Recipes)
- Enzyme solution (see Recipes)
- Suspension buffer (see Recipes)
- 0.5 M EDTA, pH 8 (see Recipes)
- 30% Ficoll (see Recipes)
- 5% Ficoll (see Recipes)
- 0.2 M Sodium phosphate, pH 8 (see Recipes)
- Lysis buffer (see Recipes)
- 1 M Mannitol (see Recipes)
- 0.2 M Sodium phosphate, pH 7.5 (see Recipes)
- Vacuole buffer (0% Ficoll) (see Recipes)
- 80% Methyl alcohol (see Recipes)
- 0.1 M Citrate (see Recipes)
- 0.2 M Sodium phosphate dibasic (see Recipes)
- Citrate phosphate buffer, pH 5.4 (see Recipes)
- Isobutyl benzene 80 µg/ml (see Recipes)
- Hexane-standard solution (see Recipes)
Equipment
- Scalpel handle #3 (Bar Naor, catalog number: BN400-3-WH )
- Beaker 400 ml (Isolab Laborgeräte, catalog number: 025.01.400 )
- Beaker 600 ml (Isolab Laborgeräte, catalog number: 025.01.600 )
- FinnpipetteTM F3 variable volume single-channel pipettes 100-1,000 µl (Thermo Fisher Scientific, catalog number: 4640060 )
- FinnpipetteTM F3 variable volume single-channel pipettes 20-200 µl (Thermo Fisher Scientific, catalog number: 4640050 )
- FinnpipetteTM F3 variable volume single-channel pipettes 2-20 µl (Thermo Fisher Scientific, catalog number: 4640030 )
- FisherbrandTM one-hole rubber stopper number 13 (Fisher Scientific, catalog number: 14-135S )
- Centrifuge 5430 R (Eppendorf, model: 5430 R , catalog number: 022620603)
- Centrifuge 5810 R (Eppendorf, model: 5810 R , catalog number: 5811000320)
- Flow hood
- Hemocytometer XB.K25 0.10 mm 1/400 mm2 (QIUJING, catalog number: 02270113 )
- Incubator at 37 °C
- Kenzan flower arrangement needle point holder 1.5 inch
- Light microscope (Olympus, model: BH-2 )
- Orbital shaker (Thermo Fisher Scientific, model: FormaTM 3250 )
- SpeedVac (Thermo Fisher Scientific, Savant, model: SVC-100H )
- Standard scissors curved 12 cm (Bar Naor, catalog number: BN11-011-12 )
- Stopcock, straight PP & HDPE, OD 8 mm (KARTELL, catalog number: 374 )
- Tissulyzer II (QIAGEN, catalog number: 85300 )
- TransferPette® S, variable 500-5,000 µl (BRAND, catalog number: 704782 )
- Tweezers curved #7 (Bar Naor, catalog number: BN-7-W )
- Tweezers extra fine #5 (Bar Naor, catalog number: BN78320-5 )
- Ultrasonic cleaner (Cole-Parmer, model: 8845-6 )
- Vacuum filter flask 2 L (Corning, catalog number: 5340-2L )
- Vacuum pump (KNF, catalog number: N 840.3 FT.18 )
- Vortex-genie 2 (Scientific industries, model: Vortex-Genie 2 , catalog number: G 560E)
Procedure
Remove all open flowers from petunia plants growing in the greenhouse 1 day before protoplast isolation (this will ensure that all open flowers are at a similar developmental stage, i.e., anthesis on the following day, Figure 1).
- Flower collection and preparation for protoplast isolation (DAY 1)
- Collect open flowers from plants growing in the greenhouse. Select completely opened flowers (at anthesis) and place them in a 600-ml beaker with 75 ml tap water. For a standard procedure, use 24 flowers which will roughly fit into four 90 x 15 mm Petri dishes at Step A7.
- All following steps must be performed in a flow hood to maintain sterility.
- Prepare 3 beakers, one containing 250 ml 5% hypochlorite and two containing 350 ml DDW. In addition, prepare four 90 x 15 mm Petri dishes containing 20 ml enzyme solution each.
- Immerse each flower using a pair of forceps in the 5% hypochlorite beaker for 30 sec and then rinse twice consecutively by immersing and shaking carefully in the DDW beakers.
- Place the sterilized flowers on clean sterile paper sheets and cut roughly 1-2 cm below the corolla (Figure 1). Remove all flower organs: stamens, flower tube, calyx, etc. (leave only the corolla). Repeat for all flowers.

Figure 1. Cutting below the corolla to remove all flower organs - Gently pierce each flower corolla using the kenzan needle point holder with the adaxial side facing up (Figure 2). Proceed immediately to the next step to prevent dehydration of the corolla.

Figure 2. Flower corolla placed on kenzan needle point holder with adaxial side facing up - Cut the flower in half and carefully place it in a Petri dish containing enzyme solution with the adaxial side facing down. Repeat this step for all flowers (approximately 12 halves fit in each dish) (Figure 3).

Figure 3. Flower halves placed in a Petri dish containing enzyme solution - Close the four Petri dishes, seal with Parafilm, cover with aluminum foil and incubate in the dark overnight (around 12 h) at 25 °C.
- Collect open flowers from plants growing in the greenhouse. Select completely opened flowers (at anthesis) and place them in a 600-ml beaker with 75 ml tap water. For a standard procedure, use 24 flowers which will roughly fit into four 90 x 15 mm Petri dishes at Step A7.
- Protoplast isolation and glycoside extraction from vacuoles (Day 2)
Protoplast isolation and vacuole isolation- Shake the covered plates for no more than 30 min at Room Temperature at 10 rpm.
- Place the 100-µm cell strainer on top of four 50-ml centrifuge tubes.
- Wet all filters with 3 ml suspension buffer.
- Gently shake the plates again by hand for a few seconds (to resuspend the cells). Using a sterile plastic 1-ml cut pipette tip, carefully collect the liquid from one Petri dish, tilt the 50-ml tube and overlay on the cell strainer. Repeat this step until all of the liquid has passed through the mesh (Figure 4). Repeat this step for the three remaining plates, collecting the liquid into the other three 50-ml tubes.
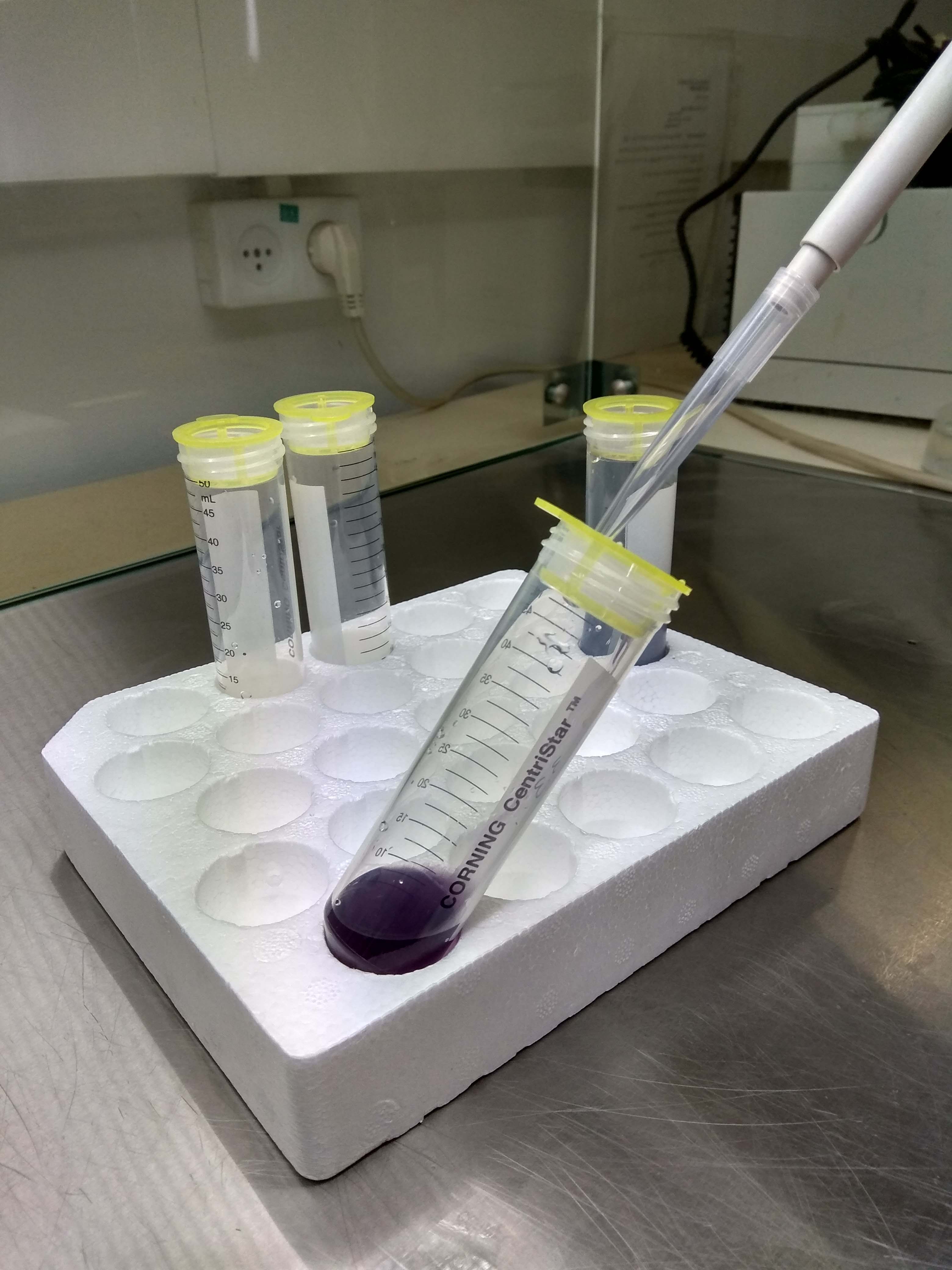
Figure 4. Collecting the protoplasts - Immediately after removing the enzyme liquid mix, use 5 ml suspension buffer to wash two dishes (this allows you to collect any protoplasts remaining in the Petri dish). Distribute the final liquid equally between the two tubes. Do the same for the other two dishes.
- Gently drip 5 ml suspension buffer onto each cell strainer to release protoplasts from the petal debris stuck on the mesh.
- Centrifuge (5810 R) the four tubes for 15 min at 100 x g at room temperature (RT) using a swing-out rotor, with acceleration set to 2 and brake (deceleration) to 0. Living protoplasts will float whereas cell debris will be pelleted (Figure 5). Keep the tube stable as movement will cause the layers to mix.
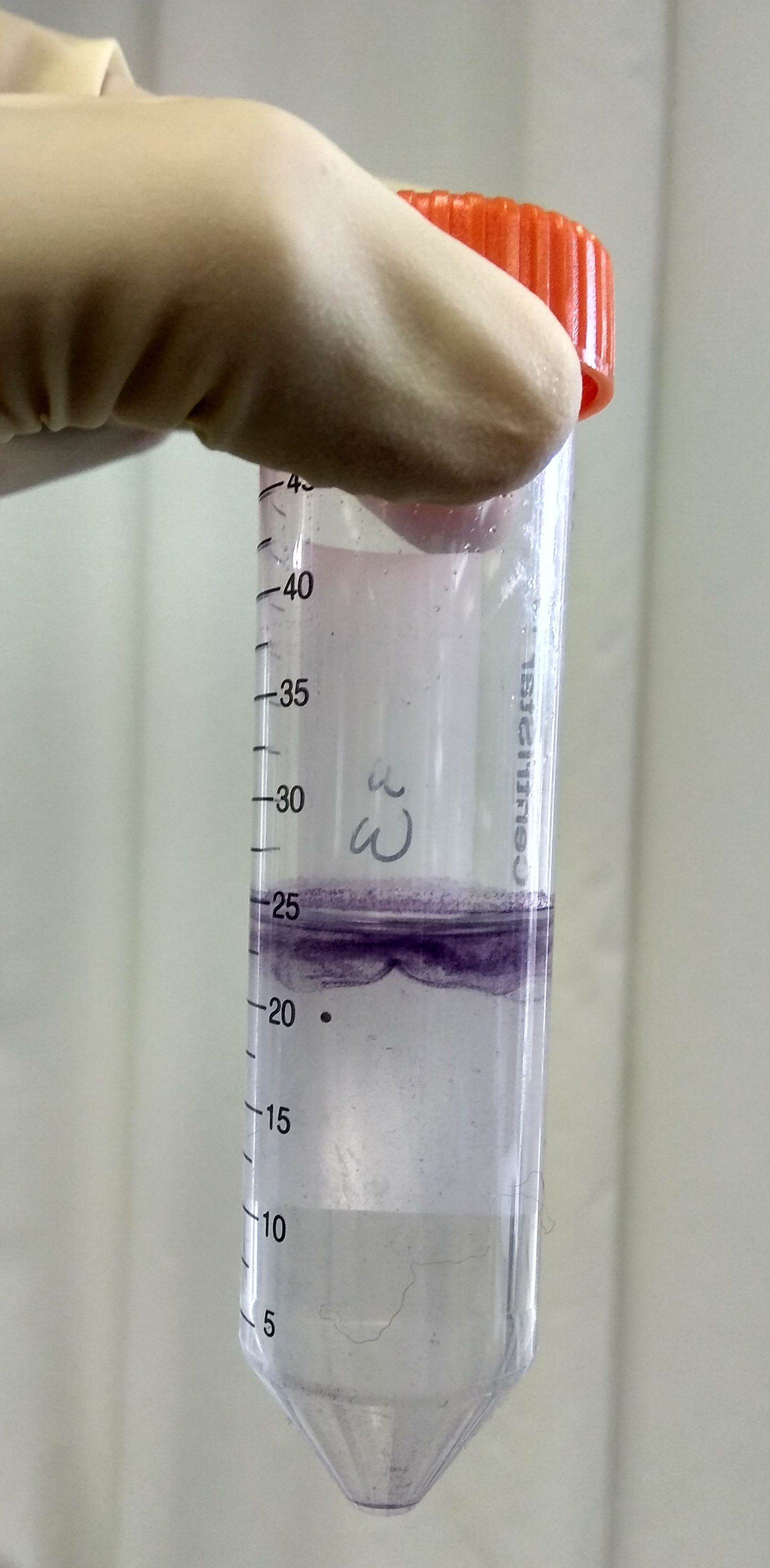
Figure 5. Purple layer with living protoplasts formed after centrifugations - Prepare the vacuum pump and vacuum filter flask with a Pasteur pipette connected to the tube (Figure 6).

Figure 6. Filter flask connected to vacuum pump - With the valve closed, start the vacuum pump and push the pipette very gently through the upper layer with the protoplasts. When the pipette has passed through the protoplast layer, slowly open the valve to initiate suction and suck up the pellet of debris and as much medium as possible but not the protoplasts (Video 1). Remember to close the valve and reduce the suction well before the protoplasts reach the bottom of the tube or your yield will be very low.Video 1. Removal of debris in the suspension solution
- Bring each tube to 25 ml with suspension buffer.
- Repeat Steps B7-B10 twice.
- Centrifuge again (4th time) and remove ca. 22 ml of debris using a Pasteur pipette, leaving 3 ml of protoplast solution. Do not leave more than 3 ml in each tube for the lysis to work.
- Count the cells under a microscope using a hemocytometer (see Data analysis; yield should be ca. 14 x 106 protoplasts/ml) (Figure 7).
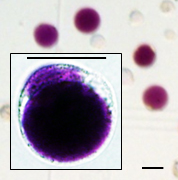
Figure 7. Light microscopy of protoplasts. Scale bar = 45 μm. - Add 10.5 ml warm (37 °C) lysis buffer to each protoplast-containing tube, pipette 5-8 times (with a cut tip) and incubate at RT for 2 min.
- Divide the contents of each tube evenly into two 15-ml tubes. Now you will have 8 tubes.
- With a 5-ml tip placed along the side of the wall and close to the surface of the liquid, add 4 ml 5% Ficoll to each of the tubes.
- As in Step B16, add 1.5 ml ice-cold vacuole buffer (0% Ficoll) to each of the tubes.
- Centrifuge all 8 tubes at 2,000 x g for 45 min at RT using a swing-out rotor, with acceleration = 2 and brake = 0.
- Vacuoles will accumulate between the 0% and 5% Ficoll fractions.
- Gently remove the upper vacuole-free Ficoll layer (transparent), and carefully transfer the vacuoles (ca. 1.5 ml) into a 2-ml Eppendorf tube with a 200-µl tip (with cut end), inserting it along the wall of the tube. Analyze vacuole intactness by light microscopy (Figure 8). Vacuoles can be stored at 4 °C for up to 1 day (see Note 6).
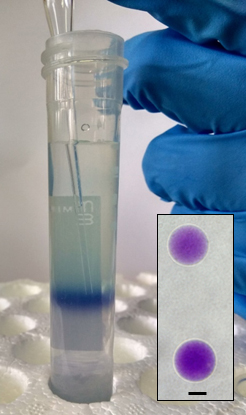
Figure 8. Ficoll fraction containing vacuoles after centrifugation. Light microscopy of intact vacuoles is shown in the inset. Scale bar = 10 μm.
Glycoside extraction from vacuoles- Transfer 0.5 ml of vacuole solution into a 2-ml Eppendorf tube (at least in triplicate to allow statistical analyses).
- Add 1.2 ml of 80% Methyl alcohol to each tube.
- Vortex vigorously for 10 sec.
- Sonicate for 20 min ( 60 Hz), vortex for 10 sec every 5 min.
- Vortex vigorously for 10 sec.
- Centrifuge (5430 R) at RT for 15 min at 16,000 x g.
- Incubate for 1 min at RT.
- Without disturbing the pellet, transfer the supernatant (ca.1.5 ml) from each tube to new 2-ml Eppendorf tubes.
- Fully evaporate (up to overnight) all of the liquid in a heated speed-vac (300 x g, 36 °C).
- Shake the covered plates for no more than 30 min at Room Temperature at 10 rpm.
- Fractionation of volatile aglycones (DAY 3)
- Add 900 µl warm (37 °C) citrate-phosphate buffer per tube with the dry pellet of extracted glycosides.
- Vortex vigorously until pellet is dissolved.
- Add 150 µl Viscozyme enzyme mixture to each tube.
- Vortex vigorously for 10 sec.
- Carefully (very slowly) add 728 µl hexane standard solution to each tube.
- Incubate statically in the dark at 37 °C for 8 h.
- Shake for 2 h at 300 rpm, RT.
- Centrifuge at 10,500 x g for 10 min at RT.
- Transfer the upper hexane phase into a 4-ml glass vial. Keep the samples at -20 °C.
- Transfer 100 µl of hexane solution into a 100-μl insert placed in a 2-ml glass vial.
- Proceed with GC-MS analyses.
- Add 900 µl warm (37 °C) citrate-phosphate buffer per tube with the dry pellet of extracted glycosides.
Data analysis
- To assay volatile aglycones in vacuoles, at least three separate experiments with different sets of 24 flowers must be performed. To establish the amounts of sequestered aglycone per vacuole, we counted vacuoles used for aglycone extraction (from separate experiments) and divided the aglycone amounts by the number of vacuoles. Standard error for the number of aglycones per vacuole was calculated with Excel.
- To assess protoplast viability, we incubated them with fluorescein diacetate solution as described in Cna’ani et al. (2017).
- To evaluate protoplast and vacuole purity, we extracted proteins and performed a western blot analysis with antibodies against the epsilon subunit of tonoplast V-type H+ATPase (V-ATPase, tonoplast), cytosolic fructose-1,6 bisphosphatase (cFBPase, cytosol), phenylalanine ammonia lyase (PAL, endomembrane/cytosol), and major light-harvesting chlorophyll a/b protein (LHCII, plastid) (Vainstein and Sharon, 1993; Howles et al., 1996; Dima et al., 2015; de Michele et al., 2016; Cna’ani et al., 2017).
- Heating and extending the evaporation time of samples prior to hydrolysis of glycosides promotes the evaporation of volatiles and is crucial to avoiding possible contamination of the vacuole fraction by aglycones from protoplasts. It is recommended that one sample extracted with hexane be analyzed prior to the hydrolysis to confirm the absence of volatile aglycones.
- To calculate the concentration of protoplasts (cells/ml), they were diluted 10 times and a 10-µl aliquot was placed on the hemocytometer counting grid without a cover glass (Figure 9A). Protoplasts were counted in 4 different squares (Figure 9B). The following formula was used: cells/ml = average number of cells per square x dilution factor x 104.

Figure 9. Protoplast concentration calculation. A. The hemocytometer used in the experiment. B. Enlargement of the counting grid. Square used for cell counting is shown in red.
Notes
- Rooted petunia plantlets (Petunia x hybrida L. line Blue Ray) were obtained from Danziger – Dan Flower Farm (Mishmar Hashiva, Israel). Plants were grown for 2 months in the greenhouse under a 25/20 °C day/night temperature regime and a 12/12 h light/dark photoperiod, with light period starting at 08:00 AM.
- This protocol was generally applied to 'Blue Ray'. Some experiments were also performed with petunia variety P720.
- On day 1, Step A8, shorter and longer incubation times will lead to low protoplast yield due to insufficient time for hydrolysis or extensive cell lysis, respectively.
- On day 2, it is highly recommended to keep the vacuum pump close to the centrifuge since the live protoplast layer tends to diffuse and resuspend, thus lowering protoplast yield.
- Use tips with cut edges for all protoplast and vacuole handling.
- It is recommended to proceed immediately to the glycoside extraction following vacuole isolation. If needed, vacuoles can be stored at 4 °C for up to 1 day.
Recipes
- TEX buffer (500 ml)
- Mix 1.55 g Gamborg B5 salts, 250 mg MES, 375 mg CaCl2, 125 mg NH4NO3, 68.46 mg sucrose
- Bring to 500 ml with distilled H2O
- Adjust pH to 5.7 with HCl
- Filter sterilize with bottle-top vacuum filter system
- Store at RT for up to 2 months
- Mix 1.55 g Gamborg B5 salts, 250 mg MES, 375 mg CaCl2, 125 mg NH4NO3, 68.46 mg sucrose
- Enzyme solution (120 ml)
- Mix 800 mg macrozyme R10 and 1.6 g cellulase R10 in a 600-ml beaker
- Bring to 120 ml with TEX buffer
- Shake for 30 min at 150 rpm
- Split into three 50-ml centrifuge tubes and centrifuge at 3,500 x g for 15 min
- Transfer supernatant to new tubes
- Make fresh
- Mix 800 mg macrozyme R10 and 1.6 g cellulase R10 in a 600-ml beaker
- Suspension buffer (500 ml)
- Mix 68.46 mg sucrose, 1.2 g HEPES, 6 g KCl, 600 mg CaCl2
- Bring to 500 ml with distilled H2O
- Adjust pH to 7.2 with HCl
- Filter sterilize with bottle-top vacuum system
- Store at RT for up to 2 months
- Mix 68.46 mg sucrose, 1.2 g HEPES, 6 g KCl, 600 mg CaCl2
- 0.5 M EDTA, pH 8 (8 ml)
- Weigh 14.89 g EDTA in a 50-ml tube
- Add 30 ml DDW
- Adjust to pH 8 with NaOH pellets
- Bring to 40 ml with DDW
- Autoclave
- Store at RT for up to 3 months
- Weigh 14.89 g EDTA in a 50-ml tube
- 30% Ficoll (50 ml)
- Weigh 15 g Ficoll in a 50-ml tube
- Bring to 50 ml with 65 °C – preheated and autoclaved DDW
- Shake at 65 °C until powder is fully dissolved (do not exceed 1 h)
- Autoclave
- Store at RT for up to 2 months
- Weigh 15 g Ficoll in a 50-ml tube
- 5% Ficoll (50 ml)
- Mix 25 ml lysis buffer and 25 ml vacuole buffer in a 50-ml tube
- Make fresh and keep at RT
- Mix 25 ml lysis buffer and 25 ml vacuole buffer in a 50-ml tube
- 0.2 M Sodium phosphate, pH 8 (10 ml)
- Weigh 138 mg NaH2PO4·H2O in a 50-ml tube. Bring to 5 ml with DDW and vortex
- Weigh 536.5 mg Na2HPO4·7H2O in a 50-ml tube. Bring to 10 ml with DDW and vortex
- Mix 0.54 ml of the NaH2PO4·H2O solution and 9.47 ml of the Na2HPO4·7H2O solution in a fresh 15-ml tube
- Store at RT for up to 3 months
- Weigh 138 mg NaH2PO4·H2O in a 50-ml tube. Bring to 5 ml with DDW and vortex
- Lysis buffer (100 ml)
- Mix 20 ml mannitol 1 M, 33.35 ml Ficoll 30%, 2 ml EDTA 0.5 M, 2.5 ml sodium phosphate 0.2 M (pH 8) in a 400 ml beaker
- Bring to 15 ml with DDW
- Make fresh and keep at 37 °C
- Mix 20 ml mannitol 1 M, 33.35 ml Ficoll 30%, 2 ml EDTA 0.5 M, 2.5 ml sodium phosphate 0.2 M (pH 8) in a 400 ml beaker
- 1 M Mannitol (50 ml)
- Weigh 9.1 g mannitol in a 300-ml beaker
- Bring to 50 ml with DDW
- Shake at 37 °C until the powder is fully dissolved
- Make 1 day ahead
- Weigh 9.1 g mannitol in a 300-ml beaker
- 0.2 M Sodium phosphate, pH 7.5 (10 ml)
- Weigh 138 mg NaH2PO4·H2O in a 50-ml tube. Bring to 5 ml with DDW and vortex
- Weigh 536.5 mg Na2HPO4·7H2O in a 50-ml tube. Bring to 10 ml with DDW and vortex
- Mix 1.6 ml of the NaH2PO4·H2O solution and 8.4 ml of the Na2HPO4·7H2O in a 15-ml tube
- Store at RT for up to 3 months
- Weigh 138 mg NaH2PO4·H2O in a 50-ml tube. Bring to 5 ml with DDW and vortex
- Vacuole buffer (0% Ficoll) (50 ml)
- Mix 12.5 ml mannitol 1 M, 1.25 ml sodium phosphate 0.2 M pH 7.5, 200 µl EDTA in a 50-ml tube
- Bring to 50 ml with DDW
- Make fresh and keep on ice
- Mix 12.5 ml mannitol 1 M, 1.25 ml sodium phosphate 0.2 M pH 7.5, 200 µl EDTA in a 50-ml tube
- Methyl alcohol 80% (40 ml)
- Mix 32 ml Methyl alcohol with 8 ml DDW in a 50-ml tube
- Make fresh
- Mix 32 ml Methyl alcohol with 8 ml DDW in a 50-ml tube
- Citrate 0.1 M (40 ml)
- Weigh 768 mg citric acid in a 50-ml tube
- Bring to 40 ml with DDW
- Make fresh
- Weigh 768 mg citric acid in a 50-ml tube
- 0.2 M Sodium phosphate dibasic (40 ml)
- Weigh 1.128 g sodium phosphate in a 50-ml tube
- Add 50 ml DDW
- Make fresh
- Weigh 1.128 g sodium phosphate in a 50-ml tube
- Citrate phosphate buffer, pH 5.4 (50 ml)
- Mix 22.2 ml of 0.1 M citric acid with 27.8 ml of 0.2 M Na2HPO4 in a 50-ml tube
- Make fresh
- Mix 22.2 ml of 0.1 M citric acid with 27.8 ml of 0.2 M Na2HPO4 in a 50-ml tube
- Isobutyl benzene (80 µg/ml)
- Take 1 µl from a stock solution, dilute in 999 µl hexane and vortex in a 1.5-ml tube
- Keep at -20 °C for up to 1 year
- Take 1 µl from a stock solution, dilute in 999 µl hexane and vortex in a 1.5-ml tube
- Hexane-standard solution (for 10 samples)
- Mix 7.2 ml hexane with 80 µl isobutyl benzene (80 µg/ml)
- Keep at -20 °C for up to 1 year
- Mix 7.2 ml hexane with 80 µl isobutyl benzene (80 µg/ml)
Acknowledgments
The presented protocol was generated based on previous works of Robert et al., 2007; Fontes et al., 2010; Faraco et al., 2011 and 2014; Pérez-Díaz et al., 2014; Shen et al., 2014 and Cna'ani et al., 2017. This work was supported by Israel Science Foundation (grant No. 247/14) and by ISF-NSFC joint research program (grant No. 2511/16). We thank Isaac Kaye for his support and assistance. A.V. is an incumbent of the Wolfson Chair in Floriculture. The authors declare that there are no conflicts of interest.
References
- Bowles, D., Isayenkova, J., Lim, E. K. and Poppenberger, B. (2005). Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8(3): 254-263.
- Cna'ani, A., Shavit, R., Ravid, J., Aravena-Calvo, J., Skaliter, O., Masci, T. and Vainstein, A. (2017). Phenylpropanoid scent compounds in Petunia x hybrida are glycosylated and accumulate in vacuoles. Front Plant Sci 8: 1898.
- Cna'ani, A., Spitzer-Rimon, B., Ravid, J., Farhi, M., Masci, T., Aravena-Calvo, J., Ovadis, M. and Vainstein, A. (2015). Two showy traits, scent emission and pigmentation, are finely coregulated by the MYB transcription factor PH4 in petunia flowers. New Phytol 208(3): 708-714.
- Dean, J. V., Mohammed, L. A. and Fitzpatrick, T. (2005). The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221(2): 287-296.
- de Michele, R., McFarlane, H. E., Parsons, H. T., Meents, M. J., Lao, J., Gonzalez Fernandez-Nino, S. M., Petzold, C. J., Frommer, W. B., Samuels, A. L. and Heazlewood, J. L. (2016). Free-flow electrophoresis of plasma membrane vesicles enriched by two-phase partitioning enhances the quality of the proteome from Arabidopsis seedlings. J Proteome Res 15(3): 900-913.
- Dima, O., Morreel, K., Vanholme, B., Kim, H., Ralph, J. and Boerjan, W. (2015). Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27(3): 695-710.
- Faraco, M., Di Sansebastiano, G. P., Spelt, K., Koes, R. E. and Quattrocchio, F. M. (2011). One protoplast is not the other! Plant Physiol 156(2): 474-478.
- Faraco, M., Spelt, C., Bliek, M., Verweij, W., Hoshino, A., Espen, L., Prinsi, B., Jaarsma, R., Tarhan, E., de Boer, A. H., Di Sansebastiano, G. P., Koes, R. and Quattrocchio, F. M. (2014). Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep 6(1): 32-43.
- Fontes, N., Silva, R., Vignault, C., Lecourieux, F., Geros, H. and Delrot, S. (2010). Purification and functional characterization of protoplasts and intact vacuoles from grape cells. BMC Res Notes 3: 19.
- Hanhineva, K., Rogachev, I., Kokko, H., Mintz-Oron, S., Venger, I., Karenlampi, S. and Aharoni, A. (2008). Non-targeted analysis of spatial metabolite composition in strawberry (Fragaria x ananassa) flowers. Phytochemistry 69(13): 2463-2481.
- Hoballah, M. E., Gubitz, T., Stuurman, J., Broger, L., Barone, M., Mandel, T., Dell'Olivo, A., Arnold, M. and Kuhlemeier, C. (2007). Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19(3): 779-790.
- Howles, P. A., Sewalt, V., Paiva, N. L., Elkind, Y., Bate, N. J., Lamb, C. and Dixon, R. A. (1996). Overexpression of L-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol 112(4): 1617-1624.
- Kortbeek, R. W., Xu, J., Ramirez, A., Spyropoulou, E., Diergaarde, P., Otten-Bruggeman, I., de Both, M., Nagel, R., Schmidt, A., Schuurink, R. C. and Bleeker, P. M. (2016). Engineering of tomato glandular trichomes for the production of specialized metabolites. Methods Enzymol 576: 305-331.
- Lashbrooke, J. G., Cohen, H., Levy-Samocha, D., Tzfadia, O., Panizel, I., Zeisler, V., Massalha, H., Stern, A., Trainotti, L., Schreiber, L., Costa, F. and Aharoni, A. (2016). MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. Plant Cell 28: 2097-2116
- Liu, J., Yang, L., Luan, M., Wang, Y., Zhang, C., Zhang, B., Shi, J., Zhao, F. G., Lan, W. and Luan, S. (2015). A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc Natl Acad Sci U S A 112(47): E6571-6578.
- Olbrich, A., Hillmer, S., Hinz, G., Oliviusson, P. and Robinson, D. G. (2007). Newly formed vacuoles in root meristems of barley and pea seedlings have characteristics of both protein storage and lytic vacuoles. Plant Physiol 145(4): 1383-1394.
- Paris, N., Stanley, C. M., Jones, R. L. and Rogers, J. C. (1996). Plant cells contain two functionally distinct vacuolar compartments. Cell 85(4): 563-572.
- Pérez-Díaz, R., Ryngajllo, M., Perez-Diaz, J., Pena-Cortes, H., Casaretto, J. A., Gonzalez-Villanueva, E. and Ruiz-Lara, S. (2014). VvMATE1 and VvMATE2 encode putative proanthocyanidin transporters expressed during berry development in Vitis vinifera L. Plant Cell Rep 33(7): 1147-1159.
- Rambla, J. L., Tikunov, Y. M., Monforte, A. J., Bovy, A. G. and Granell, A. (2014). The expanded tomato fruit volatile landscape. J Exp Bot 65(16): 4613-4623.
- Robert, S., Zouhar, J., Carter, C. and Raikhel, N. (2007). Isolation of intact vacuoles from Arabidopsis rosette leaf-derived protoplasts. Nat Protoc 2(2): 259-262.
- Schneider, S., Hulpke, S., Schulz, A., Yaron, I., Holl, J., Imlau, A., Schmitt, B., Batz, S., Wolf, S., Hedrich, R. and Sauer, N. (2012). Vacuoles release sucrose via tonoplast-localised SUC4-type transporters. Plant Biol (Stuttg) 14(2): 325-336.
- Shen, J., Fu, J., Ma, J., Wang, X., Gao, C., Zhuang, C., Wan, J. and Jiang, L. (2014). Isolation, culture, and transient transformation of plant protoplasts. Curr Protoc Cell Biol 63: 2 8 1-17.
- Shitan, N. and Yazaki, K. (2013). New insights into the transport mechanisms in plant vacuoles. Int Rev Cell Mol Biol 305: 383-433.
- Vainstein, A. and Sharon, R. (1993). Biogenesis of petunia and carnation corolla chloroplasts: changes in the abundance of nuclear and plastid-encoded photosynthesis-specific gene products during flower development. Physiol Plant 89: 192-198.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Skaliter, O., Ravid, J., Cna'ani, A., Dvir, G., Knafo, R. and Vainstein, A. (2018). Isolation of Intact Vacuoles from Petunia Petals and Extraction of Sequestered Glycosylated Phenylpropanoid Compounds. Bio-protocol 8(13): e2912. DOI: 10.21769/BioProtoc.2912.
Category
Plant Science > Plant metabolism > Metabolite profiling
Cell Biology > Organelle isolation > Vacuole
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










