- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Sleeping Beauty Transposon-based System for Rapid Generation of HBV-replicating Stable Cell Lines
Published: Vol 8, Iss 13, Jul 5, 2018 DOI: 10.21769/BioProtoc.2908 Views: 9705
Reviewed by: Yannick DebingVasudevan AchuthanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Drug Susceptibility Assay for HBV Using Southern Blotting
Sung Hyun Ahn [...] Kyun-Hwan Kim
Apr 20, 2015 11318 Views

Isolation of Exosomes from Semen for in vitro Uptake and HIV-1 Infection Assays
Marisa N. Madison [...] Chioma M. Okeoma
Apr 5, 2017 12840 Views
Abstract
The stable HBV-transfected cell lines, which based on stable integration of replication-competent HBV genome into hepatic cells, are widely used in basic research and antiviral drug evaluation against HBV. However, previous reported strategies to generate HBV-replicating cell lines, which primarily rely on random integration of exogenous DNA by plasmid transfection, are inefficient and time-consuming. We newly developed an all-in-one Sleeping Beauty transposon system (denoted pTSMP-HBV vector) for robust generation of stable HBV-replicating cell lines of different genotype. The pTSMP-HBV vector contains HBV 1.3-copy genome and dual selection markers (mCherry and puromycin resistance gene), allowing rapid enrichment of stably-transfected cells via red fluorescence-activated cell sorting and puromycin antibiotic selection. In this protocol, we described the detailed procedure for constructing the HBV-replicating stable cells and systematically evaluating HBV replication and viral protein expression profiles of these cells.
Keywords: HBVBackground
Chronic hepatitis B virus (HBV) infection is currently a major public health burden, affecting over 240 million individuals globally (Witt-Kehati et al., 2016). Patients with chronic HBV have an elevated risk of chronic active hepatitis, cirrhosis, or primary hepatocellular carcinoma (HCC) (Schweitzer et al., 2015). Current treatments with interferon-α or nucleoside analogs do not eradicate the virus, and their effects on clearing hepatitis B surface antigen (HBsAg) are limited (Lucifora and Protzer, 2016; Soriano et al., 2017). Therefore, there is an urgent need for the development of novel antiviral inhibitors (Nassal, 2015).
A cell culture model for evaluating the activity of new agents against HBV is an important tool for new drug development. The stable HBV-replicating cell lines, which carry replication-competent HBV genome stably integrated into the genome of human hepatoma cell lines (Huh7 and/or HepG2), are widely used to evaluate the effects of antiviral agents (Witt-Kehati et al., 2016). The stable HBV-producing human hepatoma cell lines (HepG2.2.15 and HepaAD38) integrated the D-genotype HBV genome, which are widely used in antiviral research (Chang et al., 1987; Ladner et al., 1997). However, stable HBV-producing cell lines of genotypes A, B, and C are not commonly used in the research field. Therefore, there is a need to develop cell lines of HBV genotypes A-C for drug development.
The Sleeping Beauty (SB) transposon system, derived from teleost fish sequences, is extremely effective at delivering DNA to vertebrate genomes, including those of humans (Structure of SB can be seen in Figure 1A). Sleeping Beauty transposition is a cut-and-paste process, during which the element ‘jumps’ from one DNA molecule to another (Figure 1B) (Ivics and Izsvak, 2011). Since its reconstruction in 1997 from the salmonid fish genome (Ivics et al., 1997), the SB system has been undergoing several modifications to improve its efficacy (Geurts et al., 2003; Baus et al., 2005; Score et al., 2006). The development of the hyperactive transposase SB100X has increased approximately 100-fold of efficiency compared with the first-generation transposase (Mátés et al., 2009), which is expected to facilitate widespread applications in functional genomics and gene therapy (Izsvak and Ivics, 2004).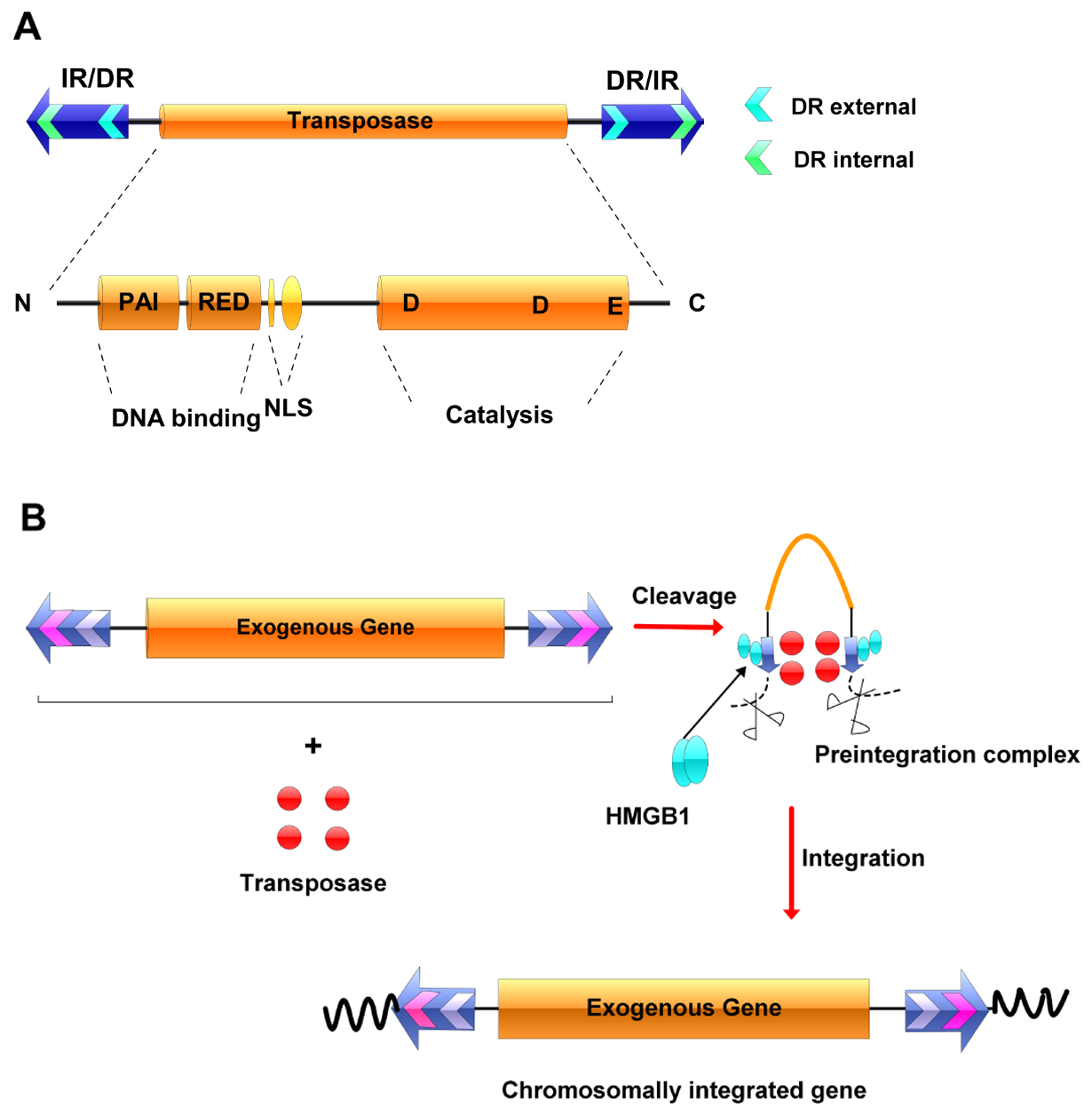
Figure 1. The Sleeping Beauty transposable element and its transposition. A. The Sleeping Beauty (SB) system. The transposase gene (yellow rectangle) is flanked by terminal inverted repeats (IR/DRs, blue arrows), each containing two binding sites for the transposase (small green arrows). The transposase consists of an N-terminal, DNA-binding domain (PAI and RED), a nuclear localization signal (NLS), a C-terminal and catalytic domain (DDE). B. Transposition. The transposase gene within the element can be replaced by a therapeutic gene, and the resultant transposon can be maintained in a simple plasmid vector. The transposase is supplied in trans. The transposase binds to its binding sites within the IR/DR repeats and, together with host factors such as HMGB1, forms a synaptic complex, in which the ends of the transposon are paired. The transposon is excised from the donor molecule and integrates into a new location.
Materials and Reagents
- Pipette tips, 10 μl (Haimen plastic, 20111088)
- Pipette tips, 200 μl (Corning, Axygen®, catalog number: T-200-Y-R )
- Pipette tips, 1 ml (Haimen plastic, 20111011)
- Cell culture plate (100 mm) (Corning, catalog number: 430167 )
- Cell culture plate (60 mm) (Thermo Fisher Scientific, catalog number: 150288 )
- 15 ml tube (Thermo Fisher Scientific, catalog number: 339651 )
- 70 μm cell strainer (Fisher Scientific, Fisherbrand, catalog number: 22-363-548 )
- Cell culture plate (6 well) (Corning, catalog number: 3516 )
- Cell culture plate (24 well) (Corning, catalog number: 3524 )
- Cell Imaging Plate, 24 well, glass bottom (Eppendorf, catalog number: 0030741021 )
- Nylon membranes (Roche Diagnostics, catalog number: 11417240001 )
- Human hepatoma HepG2 cells (Originally from the China Centre for Type Culture Collection, Wuhan, China)
- Minimum essential medium (MEM, powder) (Thermo Fisher Scientific, GibcoTM, catalog number: 41500-083 )
- Fetal bovine serum, Qualified, Australia Origin (Thermo Fisher Scientific, catalog number: 10099141 )
- X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics, catalog number: 06366236001 )
- Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31985070 )
- 0.25% Trypsin-EDTA (1x), Phenol Red (Thermo Fisher Scientific, GibcoTM, catalog number: 25200-114 )
- Puromycin (Thermo Fisher Scientific, InvitrogenTM, catalog number: A1113803 )
- Mouse anti-HBcAg (Innodx Biotechnology, catalog number: 2A7-21 [it's new anti-HBc mAb, available on request])
- Alexa Fluor® 488 Donkey Anti-Mouse IgG (H+L) (Thermo Fisher Scientific, catalog number: A-21202 )
- DAPI (Thermo Fisher Scientific, InvitrogenTM, catalog number: D1306 )
- Micrococcal nuclease (Takara Bio, catalog number: D2910 )
- Proteinase K (Takara Bio, catalog number: D9033 )
- Ethanol, C2H6O, AR (Xilong Scientific, catalog number: 1030029AR )
- Oligonucleotides
- dNTP Mixture 2.5 μM (Takara Bio, catalog number: D4030A )
- DIG Easy Hyb Granules (Roche Diagnostics, catalog number: 11796895001 )
- PrimeSTAR GXL DNA Polymerase (Takara Bio, catalog number: DR050A )
- Premix Ex TaqTM (Probe qPCR) (Takara Bio, catalog number: RR390A )
- Casein 10x blocking buffer (Sigma-Aldrich, catalog number: B6429-500ML )
- Anti-DIG (AP) antibody (Roche Diagnostics, catalog number: 11093274910 )
- CDP-Star AP substrate (Roche Diagnostics, catalog number: 12041677001 )
- Universal DNA Purification Kit (TIANGEN Biotech, catalog number: DP214-03 )
- Diagnostic kit for Hepatitis B surface antigen (CLEIA) (Wantai Biological Pharmacy, catalog number: HBV-1396 )
- Diagnostic kit for Hepatitis B e-antigen (ELISA) (Wantai Biological Pharmacy, catalog number: HBV-0396 )
- ED-11 (Innovax Biotechnology)
- Virus DNA/RNA Extraction kit (GenMag Biotechnology, catalog number: NA007 )
- Mycoplasma-free neonatal bovine serum (Tianhang Biotechnology, catalog number: 11011-8615 )
- Sodium chloride (NaCl, AR) (Xilong Scientific, catalog number: 1001012AR )
- Potassium chloride (KCl, cell culture) (Sigma-Aldrich, catalog number: P5405 )
- Hydrochloric acid (HCl, AR) (Xilong Scientific, catalog number: 1029013AR )
- Sodium hydroxide (NaOH, AR) (Xilong Scientific, catalog number: 1001037AR )
- Disodium hydrogen phosphate (Na2HPO4·12H2O, AR) (Xilong Scientific, catalog number: 1001067AR )
- Potassium dihydrogen phosphate (KH2PO4, AR) (Xilong Scientific, catalog number: 1002048AR500 )
- Sodium bicarbonate (NaHCO3, AR) (Xilong Scientific, catalog number: 44558 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: 16005-1KG-R )
- Triton X-100 (AMRESCO, catalog number: 0694 )
- Bovine albumin (Low endotoxin) (ICPbio International, catalog number: ABRE-1KG )
- Tris base (SEEBIO BIOTECH, catalog number: 183995 )
- Tris-saturated phenol (Solarbio, catalog number: T0250 )
- Ethylenediaminetetraacetic acid (EDTA, AR) (Xilong Scientific)
- NONIDET® P-40 Substitute (AMRESCO, catalog number: M158-500ML )
- Calcium chloride (CaCl2, AR) (Xilong Scientific, catalog number: U1000566-500g )
- Sodium dodecyl sulfate (SDS) (Merck, catalog number: 428015 )
- Chloroform (AR) (Xilong Scientific, catalog number: 1039013AR500 )
- Isoamyl alcohol (AR) (Xilong Scientific, catalog number: U1001975-500ml )
- Maleic acid (Sigma-Aldrich, catalog number: M0375 )
- Tween-20 (BBI Solutions, catalog number: TB0560-500ml )
- Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA-Na2·2H2O, AR) (Xilong Scientific, catalog number: 100186 )
- Acetate (CH3COOH, AR) (Xilong Scientific, catalog number: 1029047AR )
- Sodium citrate (C6H5Na3O7, AR) (Xilong Scientific, catalog number: 1001059AR )
- Verson buffer (see Recipes)
- PBS buffer (see Recipes)
- 4% paraformaldehyde (see Recipes)
- 0.2% Triton X-100 (see Recipes)
- 3% BSA (see Recipes)
- NET buffer (see Recipes)
- 1.2 M CaCl2 (see Recipes)
- 0.5 M EDTA (see Recipes)
- 10% SDS (see Recipes)
- Phenol/chloroform/isoamyl alcohol (25:24:1) (see Recipes)
- Maleic acid buffer (see Recipes)
- Washing buffer (see Recipes)
- Detection buffer (see Recipes)
- 50x TAE buffer (see Recipes)
- Depurination buffer (see Recipes)
- Denaturation buffer (see Recipes)
- Neutralization buffer (see Recipes)
- 10x SSC (see Recipes)
- 2x SSC/0.1% SDS (see Recipes)
- 0.5x SSC/0.1% SDS (see Recipes)
Equipment
- Pipettes (Mettler-Toledo International, RAININ, model: Pipet-Lite )
- CO2 Incubator (Thermo Fisher Scientific, catalog number: 3111 )
- Centrifuge (Thermo Fisher Scientific, model: HeraeusTM PicoTM 17 )
- Sorvall refrigeration Centrifuge (Thermo Fisher Scientific, model: SorvallTM ST 16R )
- BD FACS Aria III (BD, model: FACSAriaTM III )
- High Content Screening System (PerkinElmer, model: Opera PhenixTM )
- Water bath (Grant Instruments, model: GD100 )
- UV cross-linking instrument (Shanghai SIGMA High-tech, model: SH4 )
- Bio-Rad vacuum blotter (Bio-Rad Laboratories, model: Model 785 )
- Multifunctional molecular hybridization oven (UVP, model: HM-4000 )
- ImageQuant LAS4000 mini (GE Healthcare, model: ImageQuant LAS 4000 mini )
- Plastic film sealing machine (Shanghai Mingwei, model: F-400 )
- Biomek NXP (Beckman Coulter, model: Biomek NXP )
- Microplate reader (Autobio, model: PHOmo )
- Orion II Microplate Lumimometer (Titertek-Berthold, model: Orion II )
- Electrophoresis apparatus (Bio-Rad Laboratories, catalog number: 1645050 )
- LightCycler® 96 (Roche Molecular Systems, model: LightCycler® 96 )
Procedure
- Generation of HBV-replicating stable cell lines
- Transfection of HepG2 cells
- Grow HepG2 cells in MEM/10% FBS at a 37 °C, CO2 incubator. Split the cells to a 10-cm plate 24 h before transfection; the cell density should reach 70-90% at the time of DNA transfection.
- Dilute 20 μg each of pTSMP-HBV1.3 plasmids of genotype Ae (AY707087), Ba (GU357842), Ce (GU357845) and D1 (GU357846) with 2 ml opti-MEM serum-free medium, respectively. Mix gently.
Note: The map of pTSMP-HBV1.3 plasmid and the genome organization of HBV1.3 used in this study are shown in Figures 2A and 2B, respectively. The construction of pTSMP-HBV1.3 plasmids can be referred to (Wu et al., 2016).
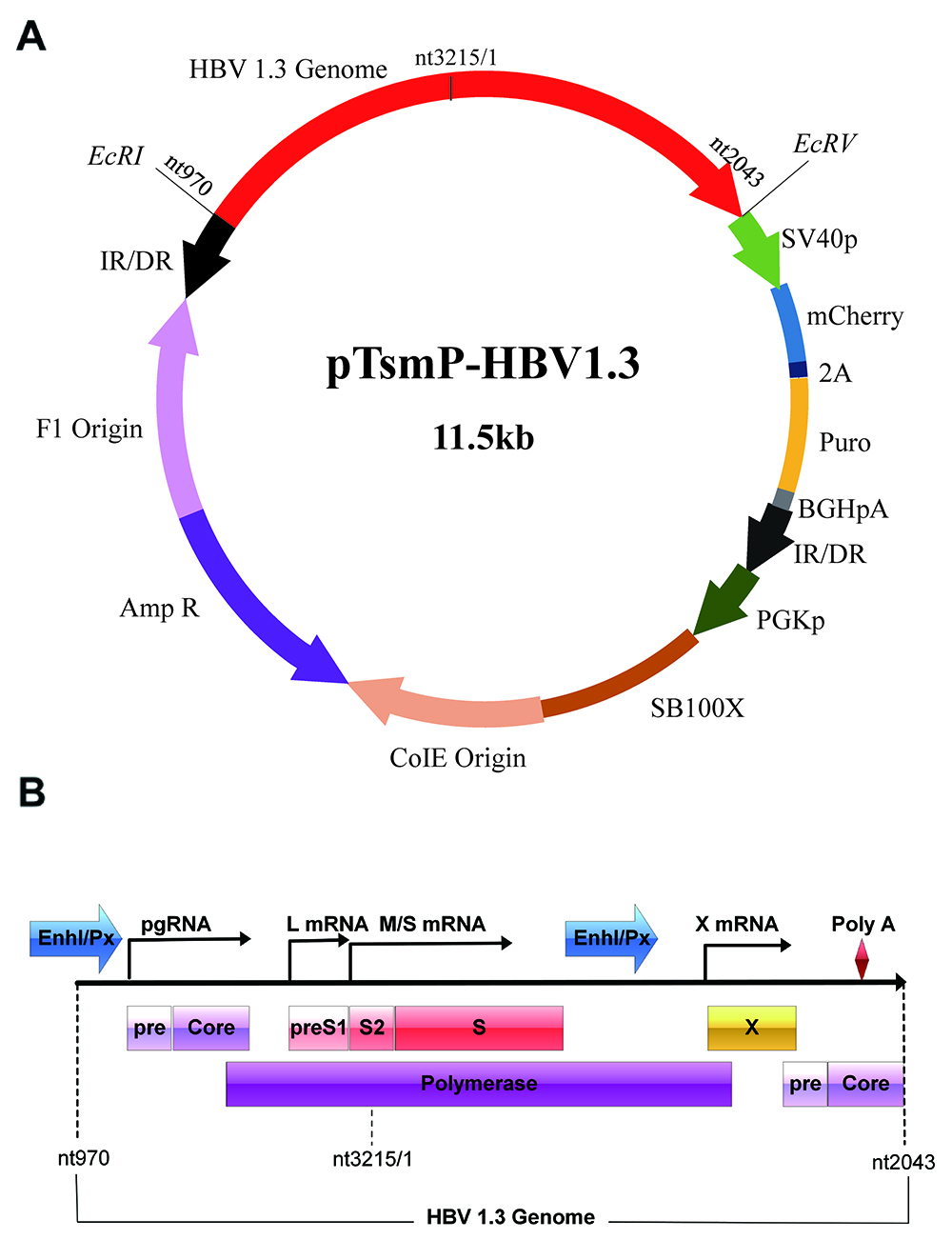
Figure 2. Schematic representation of the pTsmP vector (A) and HBV 1.3 genome construction (B)- Pipet the X-tremeGENE HP DNA Transfection Reagent (60 µl) directly into the medium containing the diluted DNA without contact with the walls of the plastic tubes. Mix gently.
- Incubate the transfection reagent: DNA complex for 15 min at room temperature.
- Add the transfection complex to the cells in a dropwise manner.
- Following transfection, incubate cells for 48 h at 37 °C in a 5% CO2 incubator; successful transfected cells with activated red fluorescence (mCherry positive) can be visualized by fluorescence microscopy and sorted by flow cytometry.
- Grow HepG2 cells in MEM/10% FBS at a 37 °C, CO2 incubator. Split the cells to a 10-cm plate 24 h before transfection; the cell density should reach 70-90% at the time of DNA transfection.
- Flow cytometric sorting of successful transfected cells
- Wash the cells with 5 ml Verson buffer (see Recipes) and discard the supernatant.
- Add 500 µl 0.25% trypsin-EDTA to the cells and incubate at 37 °C for 3 min.
- Add 5 ml MEM/10% FBS to stop digestion.
- Transfer 2 x 107 cells into a 15 ml tube.
- Centrifuge cell 1,000 x g for 3 min, and then discard supernatant.
- Wash with 5 ml PBS buffer (see Recipes).
- Centrifuge cells at 1,000 x g for 3 min, and then discard supernatant.
- Resuspend cells in 7 ml PBS buffer, and then filter through a 70 μm cell strainer.
- Run samples on BD FACS Aria III machine, the excitation light required is 561 nm (Figure 3).
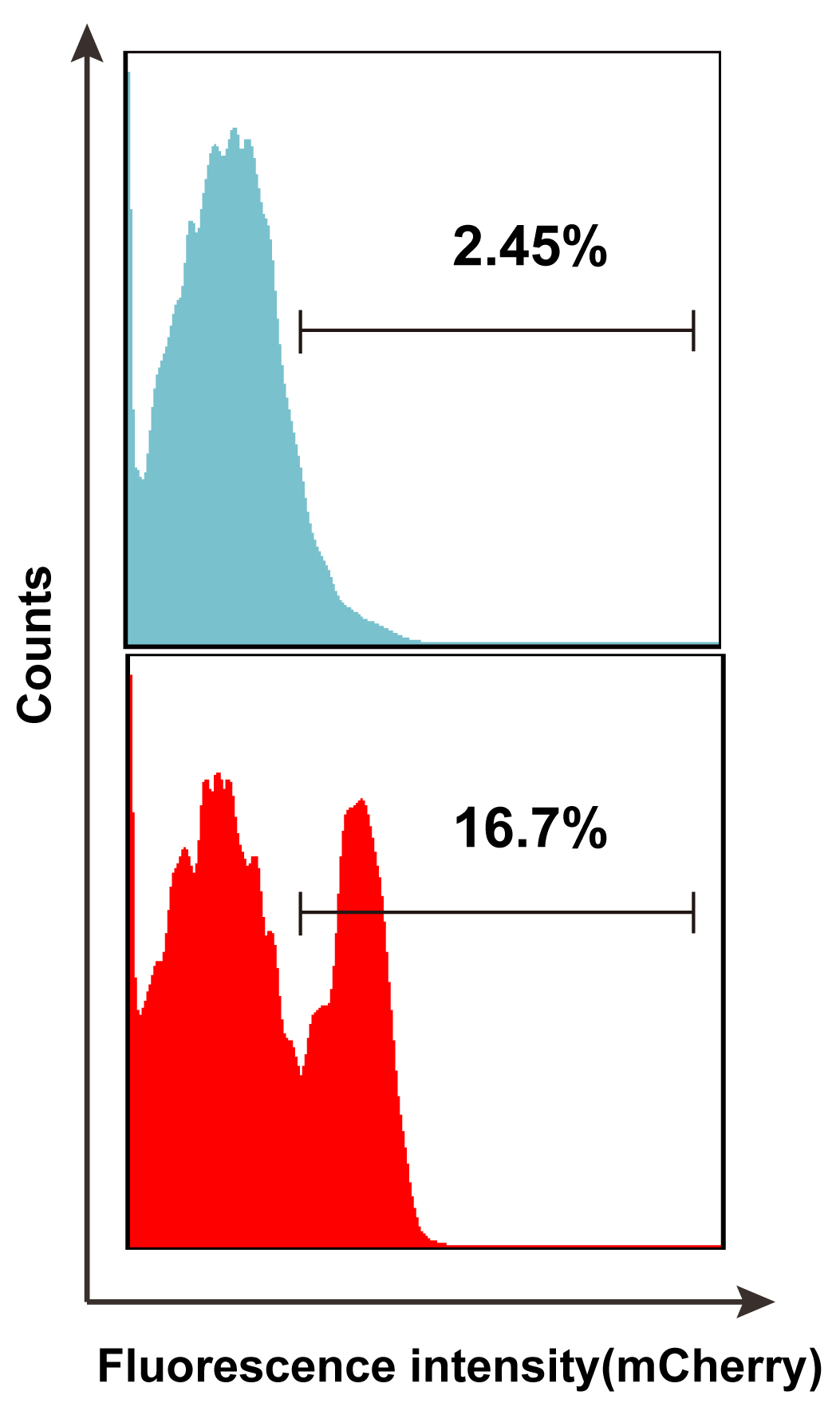
Figure 3. The first round of mCherry-activated FACS sorting. Negative cell with unexpressed mCherry is HepG2 (blue peak), positive cell with expressed mCherry is HepG2-pTsmp-1.3HBV-S11 (red peak).- The sorted mCherry-expressing cells are cultured in a 6-well culture plate for expansion in the presence of puromycin selection (2 μg/ml). Refresh Culture medium containing puromycin every 2 days.
- On the following day, when cell confluence is 70-80%, the cells are subjected to the second round of mCherry fluorescence-activated FACS sorting and further puromycin selection.
- After about a 4-round of mCherry fluorescence-activated FACS sorting and puromycin-resistant cell selection, over 90% cells are puromycin resistant and stably expressed mCherry.
- These cells are further propagated. After 10 passages, nearly 100% of cells of the 4 HBV cell lines persistently express mCherry and grow well in the presence of puromycin.
- Wash the cells with 5 ml Verson buffer (see Recipes) and discard the supernatant.
- Transfection of HepG2 cells
- Characterization of HBV-replicating stable cell lines
- Immunofluorescence of hepatitis B virus core antibody
- Cell preparation
Seed 4 x 105 pTSMP-HBV1.3 cells and 3.5 x 105 parental HepG2 cells (serve as a negative control) on the cell imaging plate with a density of 70%. Incubate the cells at 37 °C and 5% CO2 in an incubator for 24 h. - Cell washing
Discard the medium and wash once with 500 μl PBS. - Fixation
Add 250 μl per well of 4% formaldehyde (see Recipes) along the side of cell plate. Allow cells to fix in the dark for 15 min at room temperature (RT). Rinse three times in 1 ml 1x PBS for 3 min each. - Permeabilization
Incubate the samples for 5 min in 250 μl 0.2% Triton X-100 (see Recipes) per well. Rinse three times in 1 ml 1x PBS for 3 min each. - Blocking
Incubate cells with 250 μl 3% BSA (see Recipes) in PBS at RT for 1 h to block potential non-specific binding of the antibody, then discard the supernatants. - Primary antibody
Incubate the cells with 250 μl anti-HBcAg antibody (1:1,000 dilute in 3% BSA) for 1 h at RT (or overnight at 4 °C) in the dark. Next, remove the solution and wash cells 3 times with 1 ml/well PBS for 3 min each. - Secondary antibody
Incubate the cells with 250 μl secondary antibody (Alexa Fluor® 488 Donkey Anti-Mouse IgG (H+L), 1:1,000 dilute in 3% BSA) for 30 min at RT in the dark. Remove the solution and wash 3 times with 1 ml/well PBS for 3 min each in the dark. - Counterstaining
Incubate the cells with 250 μl DAPI (1:2,000 dilute in 3% BSA) for 5 min at RT in the dark. Discard the solution and wash 3 times with 1 ml/well PBS for 3 min each. - Photographs
Photograph in High Content Screening System (Figure 4). The setting used in the imager for imaging (Table 1).
Table 1. The setting used in the High Content Screening System for imaging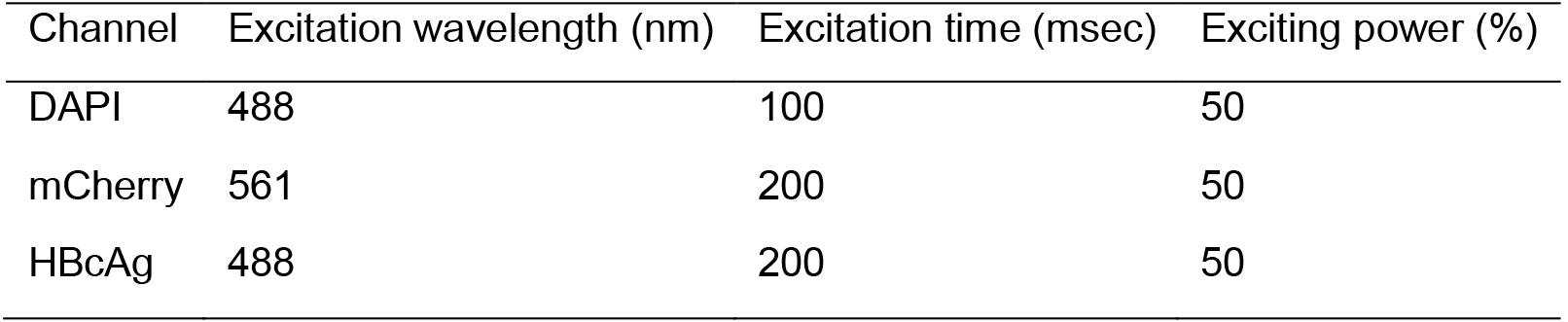
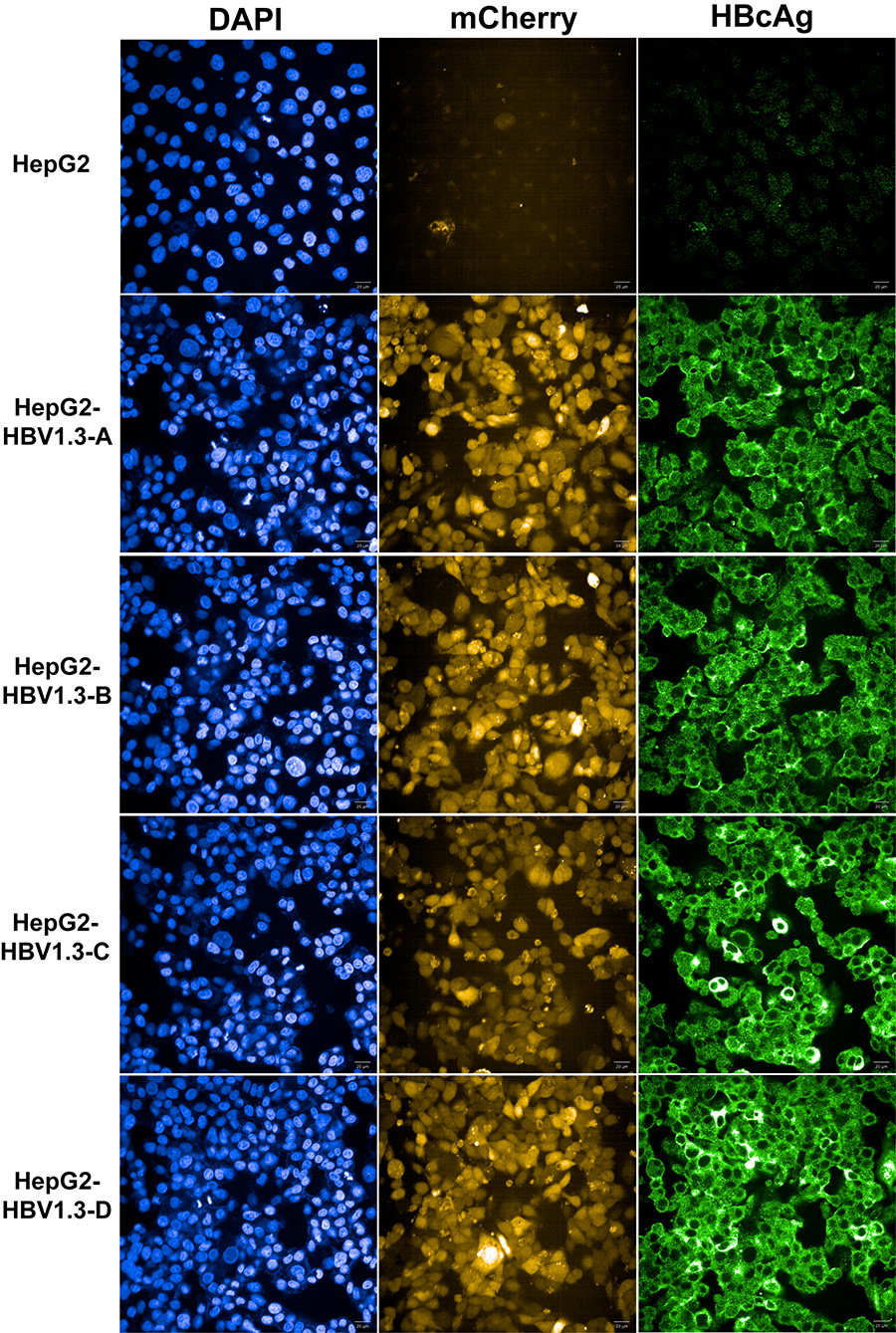
Figure 4. Immunofluorescence for HBcAg of HBV cell lines (Confocal mode, 40x water lens) - Cell preparation
- Southern blot
- Cell: Plate 4 x 106 cells in the 6 cm dish with 60-70% confluent. Culture cells for 2 days and proceed to the next experiment.
- Extraction of HBV total DNA
- Discard the supernatant of cells and wash 2 times with cold PBS.
- Add 600 μl NET buffer (see Recipes) in cell pellet and incubate on ice (or 4 °C) for 1 h.
Note: We usually add 200-300 μl in one well of a 6-well plate; or 400-600 μl a 6 cm dish; or 1.2-1.8 ml in a 10 cm dish. - Centrifuge the lysates at 13,400 x g for 20 min at 4 °C and transfer the supernatant (600 μl) into a new 1.5 ml EP tube.
- Add 2 μl Micrococcal nuclease (10 mg/ml) and 3 µl 1.2 M CaCl2 (final concentration of 6 mM) (see Recipes) and incubate for 30 min in a 37 °C water bath.
- Add 30 μl 0.5 M EDTA (final concentration of 25 mM) (see Recipes) and incubate for 15 min in a 65 °C water bath.
- Add 6.68 μl Proteinase K (19 mg/ml) and 32.1μl 10% SDS (see Recipes) and incubate overnight (At least 12 h but no more than 18 h) in a 50 °C water bath (final concentration of 200 μg/μl proteinase K and 0.5% SDS).
- Add an equal volume of phenol/chloroform/isoamylalcohol (approx. 600 μl), mix thoroughly by inverting the tube several times and incubate at room temperature (15-25 °C) for 5 min. Centrifuge at 14,000 x g for 10 min and transfer the aqueous phase into a new 1.5 ml EP tube.
- Repeat the above step (Step B2b) vii twice.
- Precipitate DNA by adding twice the volume of absolute ethanol and 1/10 volume of sodium acetate and then placing at -80 °C for 2 h. Centrifuge at 14,000 x g for 10 min at 4 °C. Carefully decant supernatant.
- Wash the DNA pellet with 500 μl 75% ethanol and centrifuge at 14,000 x g for 5 min. Carefully decant supernatant.
- Air-dry pellet for 5-10 min and re-dissolve DNA in 35 μl distilled deionized water. Store HBV DNA at -80 °C until use.
- Discard the supernatant of cells and wash 2 times with cold PBS.
- The construction of the HBV probe
- Prepare PCR reaction solution according to the following components:
5x PrimeSTAR GXL buffer 5 μl HBx-probe-F (10 μM) 1 μl HBx-probe-R (10 μM) 1 μl dNTP Mix (2.5 μM) 2 μl DIG-dUTP 2 μl pGEM-1.3 HBV 1 μl PrimeSTAR GXL DNA polymerase 1 μl ddH2O 37 μl - PCR reaction
- Purify the HBV probe by using Universal DNA Purification Kit.
- Prepare PCR reaction solution according to the following components:
- Evaluate the effect of the probe
- The probe is diluted to 2 ng/μl as the first gradient and 10-fold down-diluted to a total of 5 gradients with a minimum concentration of 0.2 pg/μl.
- Drop the diluted probe on a piece of nylon membrane via pipette, 5 μl each (final loading: 10,000 pg, 1,000 pg, 100 pg, 10 pg, 1 pg). Keep a certain distance between each point to avoid contamination.
- UV irradiation (1.5 J/cm2) for 3 min.
- Block for 30 min with 1x blocking buffer (Casein blocking buffer [10x] diluted in Maleic acid buffer [see Recipes]).
- Incubate for 40 min with anti-DIG (AP) antibody (1:3,000 diluted in 1x blocking buffer).
- Wash nylon membrane 3 times with washing buffer (see Recipes) for 10 min each.
- Rinse with detection buffer (see Recipes) for 2 min.
- Add the AP substrates to the membrane to completely infiltrate it.
- Exposure by using ImageQuant LAS4000 mini. The intensity of your experimental probe is required to make the point of 2 pg/μl or below visible to yield satisfactory hybridization results. If the probe signal is weaker than this, it will be difficult to detect signals from low abundance species (Figure 5).
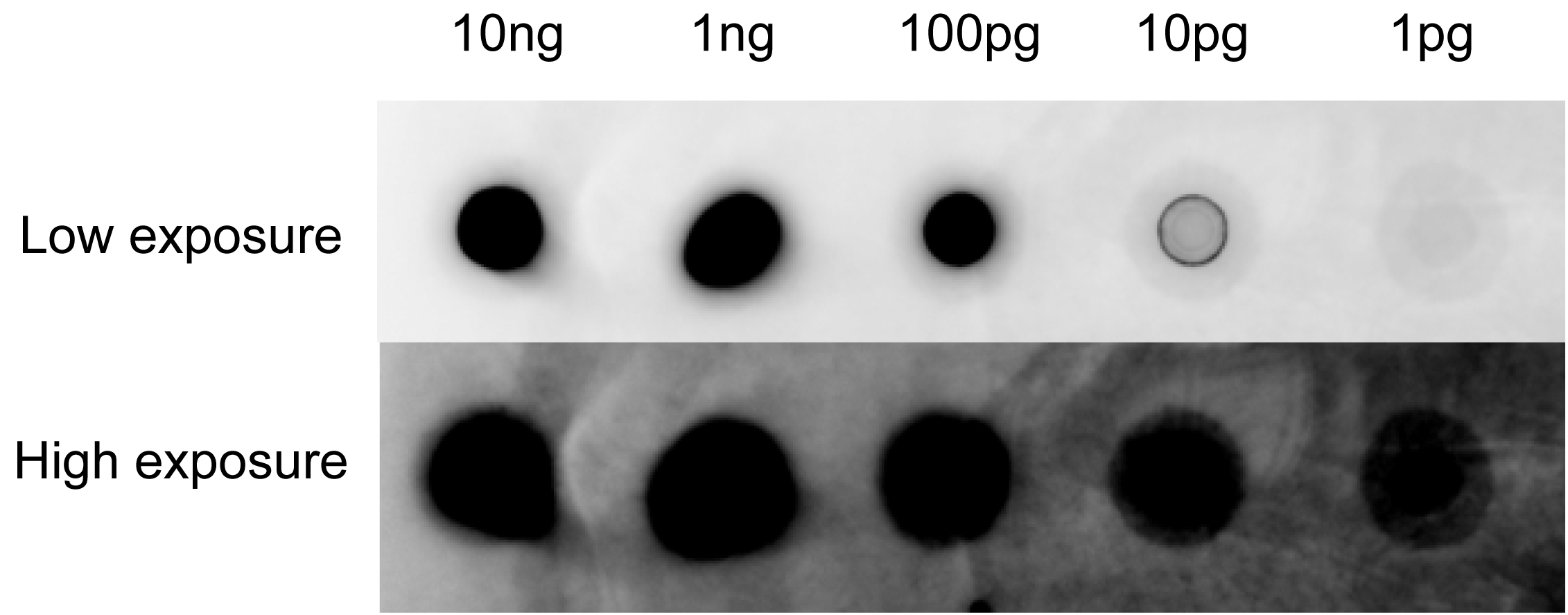
Figure 5. Southern blotting analysis for the effect of probe- Southern blot analysis
- Separate the DNA samples by electrophoresis through 1.2% agarose gel at 80 V for 2 h in 1x TAE buffer (see Recipes).
- After electrophoresis is completed, gel is treated as followed (The following steps are operated on a shaker):1)Transfer the gel to a tray and add depurination buffer (see Recipes) to soak the gel, incubate for 15 min on the shaker at RT and shake gently (Breaking the DNA into smaller pieces, thus allowing more efficient transferring from the gel to membrane).2)Remove the depurination buffer. Rinse the gel twice with ddH2O for 1 min each.3)Rinse twice with denaturation buffer (see Recipes) for 15 min each.4)Rinse the gel twice with ddH2O for 1 min each.5)Rinse with neutralization buffer (see Recipes) for 10 min.
- Transfer DNA from the gel to nylon membrane with 10x SSC for 1.5 h by vacuum blotter (Figure 6).
Note: Wet the precut nylon membrane in double distilled water by slowly lowering the membrane at a 45-degree angle to the water. Then, wet the membrane and the filter paper in 10x SSC; Place the gel and nylon membrane in the order shown in Figure 6; Remove bubbles between the gel and membrane; Start the vacuum source and slowly turn the bleeder valve clockwise until the gauge reads at 5 inches of Hg.
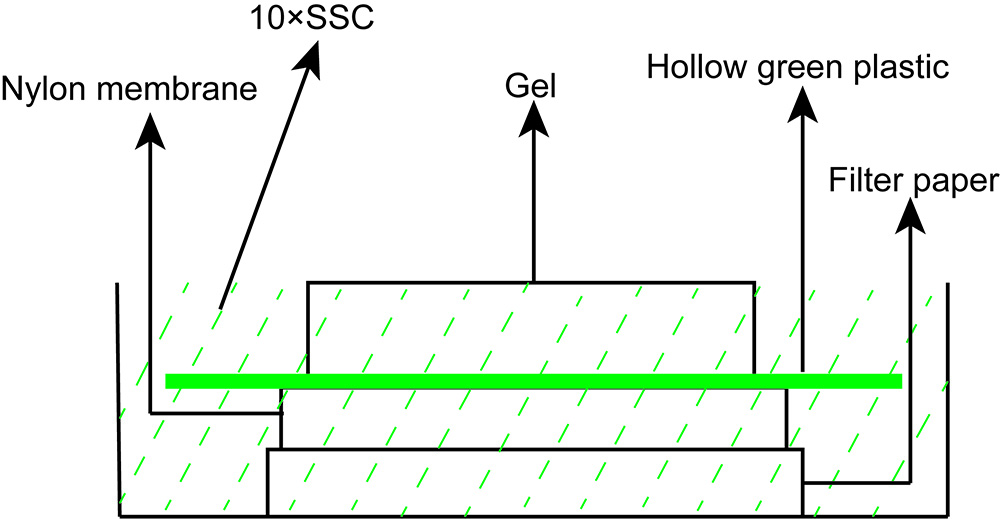
Figure 6. Scheme of capillary transfer method for Southern blotting- After gel transfer, fix the DNA to the nylon membranes by UV crosslinking at 1.5 J/cm2 for 3 min.
Note: Irradiation time = irradiation dose (mJ/cm2)/irradiation intensity (mW/cm2), the irradiation intensity of this instrument is 9.0 mW/cm2. - Pre-hybridization step
Place membrane in hybridization plastic film contained with 10 ml DIG Easy Hyb buffer and incubate the membrane for 2 h in 42 °C hybridization chamber with gentle agitation. - Hybridization1)Denature the DNA probe at 95 °C for 5 min and immediately place on ice for 2 min.2)Replace pre-warmed 10 ml of DIG Easy Hyb buffer with 300 ng DIG-dUTP- labeled HBV probe and incubate overnight in a 42 °C hybridization chamber with gentle agitation.
- Wash1)Wash 2 times with 2x SSC/0.1% SDS buffer for 5 min at room temperature.2)Wash 2 times with pre-warmed 0.5x SSC/0.1% SDS buffer for 5 min at 65 °C.
Note: Shake the buffer before use. - Detection1)Rinse with Maleic acid buffer (see Recipes) for 2 min.2)Block for 30 min with 1x blocking buffer (10x blocking buffer diluted with Maleic acid buffer).3)Incubate for 40 min with anti-DIG(AP) antibody (1:3,000 diluted in 1x blocking buffer)4)Wash 3 times with 100 ml washing buffer for 15 min each.
Note: Washing buffer shake evenly before use.5)Equilibrate for 3 min in 20 ml detection buffer.6)Add the membrane with AP substrate in dropwise so that to completely infiltrate.7)Expose the membrane continuously and take images using ImageQuant LAS4000 mini.
- Separate the DNA samples by electrophoresis through 1.2% agarose gel at 80 V for 2 h in 1x TAE buffer (see Recipes).
- Detection of the levels of viral markers in supernatants
- Cell
- Seed 3.5 x 105 pTSMP-HBV1.3 cells in the 24-well plates with a density of 60-70%.
- Replenish and collect the culture medium for measurement of viral antigens and HBV DNA every 2 days.
- Monitor the levels of these viral markers in supernatants for each cell line during 1 month.
- Seed 3.5 x 105 pTSMP-HBV1.3 cells in the 24-well plates with a density of 60-70%.
- Viral antigens
- Measure viral antigens (HBsAg and HBeAg) in culture medium by chemiluminescence method using commercial assay kits (Wantai, Beijing, China).
- Antigen (HBsAg and HBeAg) standards are standardized patient serum, with the original concentration are 90,000 IU/ml and 10,000 Ncu/ml respectively.
- The standard of HBeAg is diluted with 20% NBS into six gradient concentrations, 10, 5, 2.5, 1.25, 0.25, 0.05 Ncu/ml respectively.
- The standard of HBsAg is diluted with ED-11 into six gradient concentrations, 45, 9, 1.8, 0.36, 0.072, 0.0144 IU/ml respectively.
- Measure viral antigens (HBsAg and HBeAg) in culture medium by chemiluminescence method using commercial assay kits (Wantai, Beijing, China).
- HBV total DNA
- HBV total DNA extraction1)Extract the total DNA in culture medium by using virus DNA/RNA extraction kit on Biomek NXP (Beckman, California, USA).2)The standard of HBV total DNA is standardized patient serum with the original concentration is 1.5 x 108 IU/ml, diluting with 10-fold dilutions in PBS into six gradients.
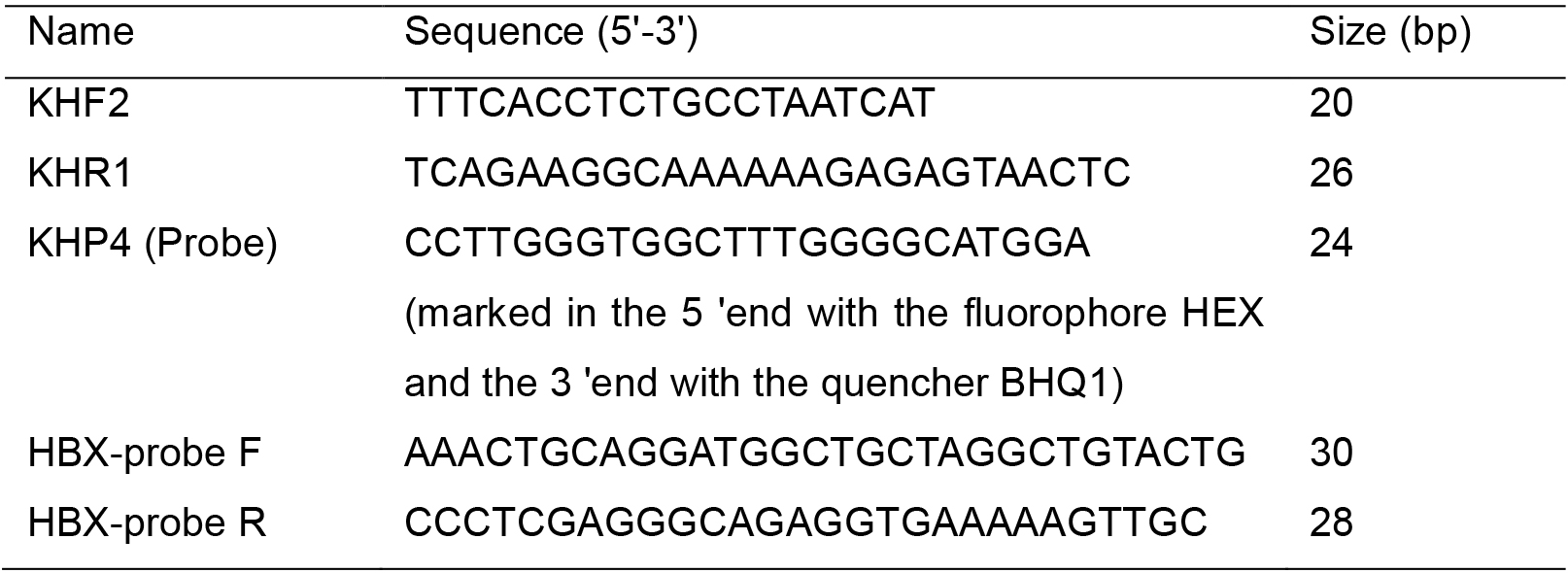
- Prepare PCR reaction solution according to the following components.
2x Premix Ex Taq (Probe qPCR) 10 μl KHF2 (100 μM) 0.1 μl KHR1 (100 μM) 0.1 μl KHP4 (100 μM) 0.05 μl DEPC water 4.75 μl Template 5 μl - Real-time PCR reaction is performed with LightCycler 96.
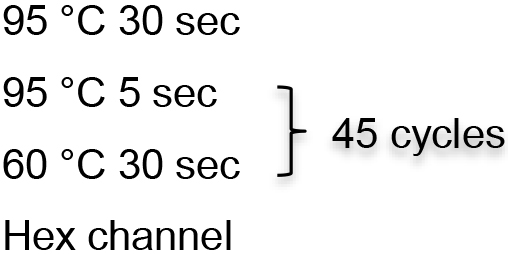
- HBV total DNA extraction
- Cell
- The probe is diluted to 2 ng/μl as the first gradient and 10-fold down-diluted to a total of 5 gradients with a minimum concentration of 0.2 pg/μl.
Notes
- The state of the cells is critical to the transfection efficiency and cell state after sorting.
- During the sorting process, cells should be sorted at 4 °C, and the flow rate should be controlled below 9 µl/min. This is crucial for the cell state after sorting.
- The number of cells after sorting should at least be enough to plate in 48-well plates so that the cells are more likely to survive.
Recipes
- Verson buffer(1 L)
0.4 g KCl
0.06 g KH2PO4
8.0 g NaCl
0.35 g NaHCO3
0.08 g Na2HPO4·12H2O
0.2 g EDTA
Filter sterilize (0.22 μm) or sterilize by autoclavation - PBS buffer
145 mmol/L NaCl
2.68 mmol/L KCl
10 mmol/L Na2HPO4·12H2O
1.76 mmol/L KH2PO4
Adjust to pH 7.4
Filter sterilize (0.22 μm) or sterilize by autoclavation - 4% Paraformaldehyde
4 g Paraformaldehyde
100 ml PBS
1-2 drops 10 M NaOH
Store at 4 °C and protect from light - 0.2% Triton X-100
200 μl Triton X-100
100 ml PBS
Store at 4 °C - 3% BSA
3 g BSA
100 ml PBS
Sub-package and store at -20 °C - NET buffer
50 mM Tris-base (pH = 8.0)
1 mM EDTA
100 mM NaCl
0.5% NP-40 - 1.2 M CaCl2
13.32 g CaCl2
100 ml ultra-pure water
Store at 4 °C - 0.5 M EDTA
Store at 4 °C - 10% SDS
100 g electrophoresis grade SDS
Add ddH2O to 1 L
Heat to 68 °C
Adjust to pH 7.2 - Phenol/chloroform/isoamyl alcohol (25:24:1)
Equilibrate phenol, chloroform and isoamyl alcohol in a ratio of 25:24:1, then store in a brown glass bottle and cover with a layer of Tris-Cl, store at 4 °C until use - Maleic acid buffer
0.1 M Maleic acid
0.15 M NaCl
Adjust to pH 7.5
Sterilize by autoclavation - Washing buffer
0.1 M Maleic acid
0.15 M NaCl
3 ml/L Tween 20
Adjust to pH 7.5
Sterilize by autoclavation - Detection buffer
0.1 M Tris-base
0.1 M NaCl
Adjust to pH 9.5
Sterilize by autoclavation - 50x TAE buffer
242 g Tris-base
100 ml EDTA (0.5 M, pH 8.0)
57.1 ml acetate
Add ddH2O to 1 L
Adjust to pH 8.5 - Depurination buffer
0.2 N HCl
Sterilize by autoclavation - Denaturation buffer
0.5 M NaOH
1.5 M NaCl
Sterilize by autoclavation
Neutralization buffer
1 M Tris-base
1.5 M NaCl
Sterilize by autoclavation - 10x SSC
1.5 M NaCl
0.15 M Sodium citrate
Adjust to pH 7.0
Sterilize by autoclavation - 2x SSC/0.1% SDS
200 ml 10x SSC
10 ml 10% SDS
Add ultra-pure water to 1 L
Sterilize by autoclavation - 0.5x SSC/0.1% SDS
50 ml 10x SSC
10 ml 10% SDS
Add ultra-pure water to 1 L
Sterilize by autoclavation
Acknowledgments
We would like to acknowledge the following publication on which this protocol is based: Wu et al. (2016). The authors declare no competing financial interests.
References
- Baus, J., Liu, L., Heggestad, A. D., Sanz, S. and Fletcher, B. S. (2005). Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther 12(6): 1148-1156.
- Chang, C. M., Jeng, K. S., Hu, C. P., Lo, S. J., Su, T. S., Ting, L. P., Chou, C. K., Han, S. H., Pfaff, E., Salfeld, J. and et al. (1987). Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J 6(3): 675-680.
- Geurts, A. M., Yang, Y., Clark, K. J., Liu, G., Cui, Z., Dupuy, A. J., Bell, J. B., Largaespada, D. A. and Hackett, P. B. (2003). Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther 8(1): 108-117.
- Ivics, Z. and Izsvak, Z. (2011). Nonviral gene delivery with the Sleeping Beauty transposon system. Hum Gene Ther 22(9): 1043-1051.
- Ivics, Z., Hackett, P. B., Plasterk, R. H. and Izsvak, Z. (1997). Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91(4): 501-510.
- Izsvak, Z. and Ivics, Z. (2004). Sleeping beauty transposition: biology and applications formolecular therapy. Mol Ther 9(2): 147-156.
- Ladner, S. K., Otto, M. J., Barker, C. S., Zaifert, K., Wang, G. H., Guo, J. T., Seeger, C. and King, R. W. (1997). Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 41(8): 1715-1720.
- Lucifora, J. and Protzer, U. (2016). Attacking hepatitis B virus cccDNA--The holy grail to hepatitis B cure. J Hepatol 64(1 Suppl): S41-S48.
- Mátés, L., Chuah, M. K., Belay, E., Jerchow, B., Manoj, N., Acosta-Sanchez, A., Grzela, D. P., Schmitt, A., Becker, K., Matrai, J., Ma, L., Samara-Kuko, E., Gysemans, C., Pryputniewicz, D., Miskey, C., Fletcher, B., VandenDriessche, T., Ivics, Z. and Izsvak, Z. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41(6): 753-761.
- Nassal, M. (2015). HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64(12): 1972-1984.
- Schweitzer, A., Horn, J., Mikolajczyk, R. T., Krause, G. and Ott, J. J. (2015). Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386(10003): 1546-1555.
- Score, P. R., Belur, L. R., Frandsen, J. L., Geurts, J. L., Yamaguchi, T., Somia, N. V., Hackett, P. B., Largaespada, D. A. and McIvor, R. S. (2006). Sleeping Beauty-mediated transposition and long-term expression in vivo: use of the LoxP/Cre recombinase system to distinguish transposition-specific expression. Mol Ther 13(3): 617-624.
- Soriano, V., Barreiro, P., Benitez, L., Pena, J. M. and de Mendoza, C. (2017). New antivirals for the treatment of chronic hepatitis B. Expert Opin Investig Drugs 26(7): 843-851.
- Witt-Kehati, D., Bitton Alaluf, M. and Shlomai, A. (2016). Advances and challenges in studying hepatitis B virus in vitro. Viruses 8(1).
- Wu, Y., Zhang, T. Y., Fang, L. L., Chen, Z. X., Song, L. W., Cao, J. L., Yang, L., Yuan, Q. and Xia, N. S. (2016). Sleeping Beauty transposon-based system for rapid generation of HBV-replicating stable cell lines. J Virol Methods 234: 96-100.
- Cell: Plate 4 x 106 cells in the 6 cm dish with 60-70% confluent. Culture cells for 2 days and proceed to the next experiment.
- Immunofluorescence of hepatitis B virus core antibody
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zheng, J., Cao, J. and Yuan, Q. (2018). Sleeping Beauty Transposon-based System for Rapid Generation of HBV-replicating Stable Cell Lines. Bio-protocol 8(13): e2908. DOI: 10.21769/BioProtoc.2908.
Category
Microbiology > Microbe-host interactions > In vitro model > Cell line
Microbiology > Microbe-host interactions > Virus
Cell Biology > Cell structure > Chromosome
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











