- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Membrane Protein Interactions by Cell-based Tango Assays
Published: Vol 7, Iss 22, Nov 20, 2017 DOI: 10.21769/BioProtoc.2903 Views: 10851
Reviewed by: Alexandros AlexandratosLokesh KalekarAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Cell-Sonar, an Easy and Low-cost Method to Track a Target Protein by Expression Changes of Specific Protein Markers
Sabrina Brockmöller [...] Simone Rothmiller
Feb 5, 2025 1657 Views

Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells
Urbi Mukhopadhyay [...] Sagar Bhogaraju
Jun 20, 2025 2435 Views

Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1207 Views
Abstract
The Tango assay is a protein-protein interaction assay, in which a transcription factor (rTA) is fused to a membrane-bound protein via a linker that contains a cleavage site for TEV protease, whereas a soluble interaction partner is fused to TEV protease (Barnea et al., 2008). Association between the two interaction partners leads to an efficient cleavage of the transcription factor, allowing it to translocate to the nucleus and activate a luciferase reporter gene as measurement of the interactions. In this modified assay, we fused one copy of the membrane-spanning amyloid precursor protein (APP) C99 region to TEV site-rTA (C99-TEV site-rTA) and a second copy to TEV protease (C99-TEV) to analyze intramembrane C99-C99 interaction in live cells.
Keywords: Tango assayBackground
The amyloid precursor protein (APP) has three dimerization domains in its N-terminal extracellular domain. In addition, APP can also form dimers through the membrane-bound C99 (C-terminal 99 amino acid fragment) region. Importantly, C99 dimerization has been linked to Aβ production in Alzheimer’s disease (AD) pathology. The Tango assay described here and schematically shown as cartoon in Figure 1 is a fast and sensitive method for investigating homodimerization of C99 and other membrane proteins (Yan et al., 2017).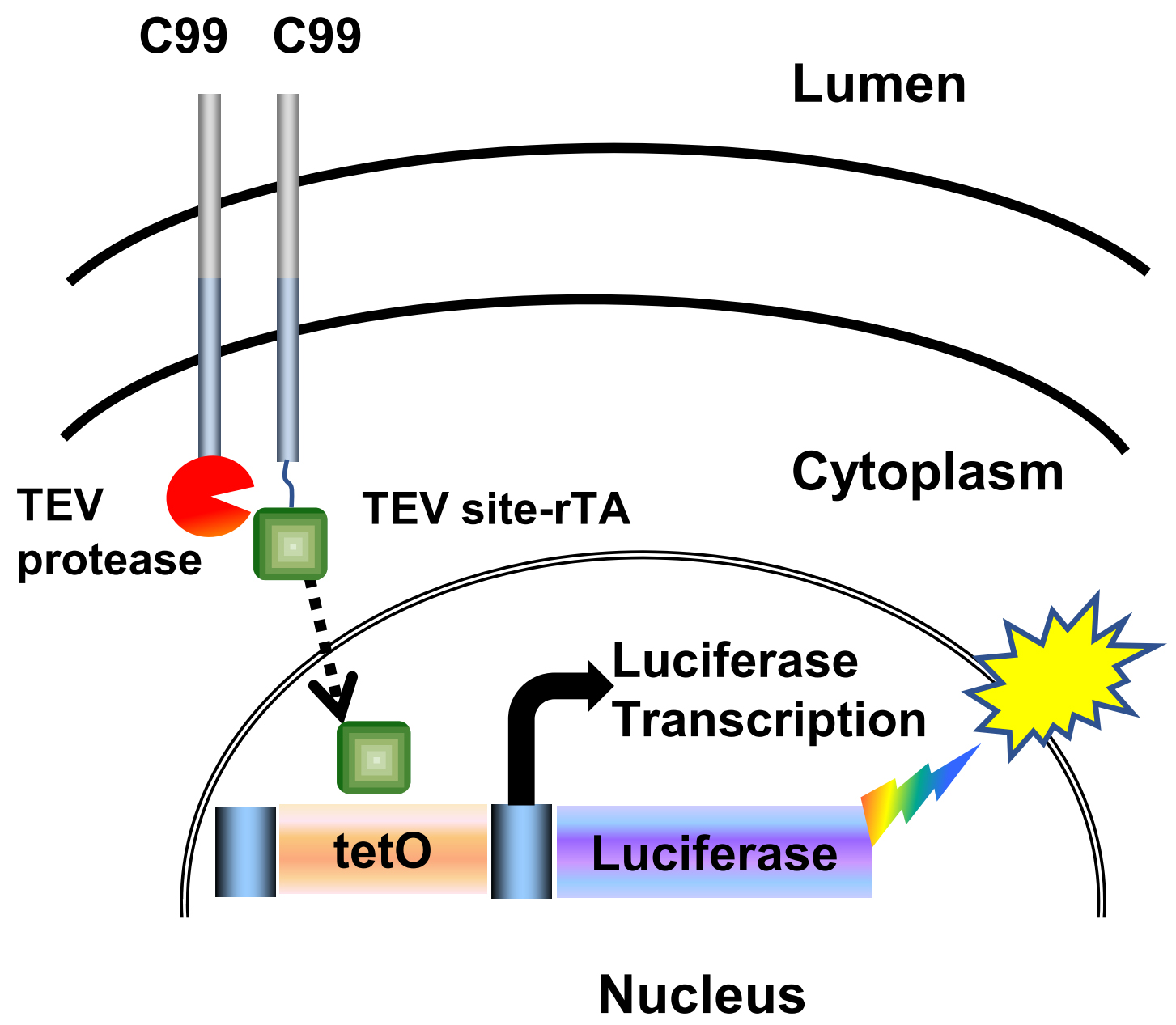
Figure 1. Cartoon illustration of the Tango interaction assay. Upon membrane cleavage of the C99 hybrid protein by TEV protease, the rTA transactivator protein is released from the membrane into the cytoplasm. This allows rTA to enter the nucleus and bind the tetO DNA-binding site upstream of an integrated luciferase reporter gene to stimulate luciferase reporter gene activity as measured by luminescence. (Yan et al., 2017).
Here we use the Dual-luciferase reporter assay kit. The stably integrated luciferase-Firefly reads represent the γ-secretase cleavage activity, while the transfected Renilla luciferase reads serve as normalization standard.
Materials and Reagents
- Pipette tips (VWR)
- 24-well plate (Corning, Costar®, catalog number: 3524 )
- 96-well plate (Corning, catalog number: 3595 )
- Cell culture flask (Corning, catalog number: 430639 )
- 96-well OptiPlate (PerkinElmer, catalog number: 6005290 )
- Gel loading pipette tip
- PS1/PS2-deleted HTL cells (PS1/PS2 gene were deleted by CRISPR/Cas9 from HTL cells [Xu et al., 2016])
- Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 11965092 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 26140079 )
- Trypsin 0.25%-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056 )
- Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31985062 )
- Dual-luciferase reporter assay kit (Promega, catalog number: E1960 )
- X-tremeGENE 9 Reagent (Roche Diagnostics, catalog number: 06365787001 )
- Sodium phosphate dibasic (Na2HPO4) (Fisher Scientific, catalog number: S374-1 )
- Potassium phosphate monobasic (KH2PO4) (EMD Millipore, catalog number: PX1565-1 )
- Sodium chloride (NaCl) (EMD Millipore, catalog number: SX0420-5 )
- Potassium chloride (KCl) (Fisher Scientific, catalog number: BP366-500 )
- 10x phosphate buffered saline (PBS buffer) (see Recipes)
- 1x passive lysis buffer (PLB) (see Recipes)
- LAR2 substrate (see Recipes)
- Stop & Glo® Reagent (see Recipes)
Equipment
- Micro pipette (Eppendorf)
- Vacuum pump
- 37 °C, 5% CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM Series II 3110 Water-Jacketed)
- Cell culture microscope (Nikon Instruments, model: Eclipse TS100 )
- Shaker (ARMA LAB, model: Orbital Shaker 100 )
- Biosafety cabinet (The Baker, model: SterilGARD® e3 )
- EnVision Multilabel Plate Reader (PerkinElmer, model: EnVision Multilabel )
Software
- GraphPad Prism 5 (https://www.graphpad.com/scientific-software/prism/)
Procedure
- Cell culture
- The PS1/PS2-deleted HTL cells are routinely grown in DMEM supplemented with 10% (v/v) FBS at 37 °C under a humidified 5% CO2 atmosphere.
- Treat the cells with 0.25% trypsin-EDTA at 37 °C for about 2 min. Dilute the cells to 0.4 x 106/ml with DMEM medium (supplemented with 10% [v/v] fetal bovine serum) and split at 50,000 per well into a 24-well plate one day prior to transfection.
- The PS1/PS2-deleted HTL cells are routinely grown in DMEM supplemented with 10% (v/v) FBS at 37 °C under a humidified 5% CO2 atmosphere.
- Transfection
- Prepare a master mix for one sample:
To a sterile 1.5 ml Eppendorf tube, add:
10 µl Opti-MEM medium
0.195 µl X-tremeGENE 9 Transfection Reagent.
Notes:- Avoid touching the side of the tube while adding reagent.
- Make a master mix for larger sample numbers: for example, for 10 samples, multiply by 11 to prepare some extra mix. (10 x 11 = 110 µl medium; 0.195 x 11 = 2.145 µl X-tremeGENE 9 Transfection Reagent).
- Avoid touching the side of the tube while adding reagent.
- Add 10 µl Opti-MEM medium per well in a 96-well plate and then add 65 ng total DNA into each well.
Note: We usually dilute the DNA plasmids to a concentration of 100 ng/µl and store them at 4 °C. - Add 10 µl master mix to DNA mix in each well of the 96-well plate and mix gently by pipetting.
- Incubate at room temperature for about 30 min.
- Carefully transfer 20 µl mix by pipetting into each well of cells cultured in a 24-well plate.
Note: In this assay, 10 ng C99 (or its variants)-TEV site-rTA expression construct, 10 ng C99 (or its variants)-TEV protease construct and 5 ng of phRG-tk Renilla normalization luciferase expression vector were transfected together with 40 ng pBSK plasmid control into PS1/PS2-deleted HTL cells. - Culture the cells in DMEM supplemented with 10% (v/v) fetal bovine serum at 37 °C under a humidified 5% CO2 atmosphere.
- Prepare a master mix for one sample:
- Luciferase measurement
- One day post transfection, remove the medium from the cultured cells by vacuum pump attached by tubing to a liquid trap and a gel loading pipette tip, and gently apply a sufficient volume (i.e., 500 µl/well) of PBS (see Recipes) to rinse the bottom of each well.
- Dispense 100 µl of 1x PLB (see Recipes) into each well and gently shake the culture plate for 15 min at room temperature (e.g., 70 rpm).
Note: We use Eppendorf® Research® Pro electronic single channel pipette (20-1,000 µl) to dispense the PLB buffer. However, any laboratory pipette that can be set to 100 µl can be used. - Transfer 20 µl PLB lysate without cell debris to each well of a 96-well OptiPlate.
Note: Generally, it is unnecessary to clear lysates of residual cell debris prior to performing the assay. - Program the EnVision Multilabel Plate Reader to perform a 2-sec premeasurement delay, followed by a 10-sec measurement period using the Envision 96 Plate US Luminescence (700 nm emission filter) aperture for each reporter assay.
- Dispense 50 µl LAR2 substrate (see Recipes) into each well of the 96-well OptiPlate, mix by pipetting 2 or 3 times and measure Firefly luciferase activity using EnVision Multilabel Plate Reader. The Luminescence reads represent the Firefly luciferase activity.
- Dispense 50 µl Stop & Glo® Reagent (see Recipes) into the same plate, mix by pipetting 2 or 3 times and measure the Renilla luciferase activity by EnVision Multilabel Plate Reader. The Luminescence reads represent the Renilla luciferase activity.
- One day post transfection, remove the medium from the cultured cells by vacuum pump attached by tubing to a liquid trap and a gel loading pipette tip, and gently apply a sufficient volume (i.e., 500 µl/well) of PBS (see Recipes) to rinse the bottom of each well.
Data analysis
Each data point represents the average from at least three independent experiments (Table 1), calculated activities are shown in Table 2. Data are presented as grouped bar graph type. The statistical analysis of data can be accomplished with GraphPad Prism using the two-tailed Student’s t-test versus control (Figure 2). A practical example of the applicability of this Tango interaction assay was recently shown in (Yan et al., 2017).
Table 1. Original Firefly and Renilla activity reads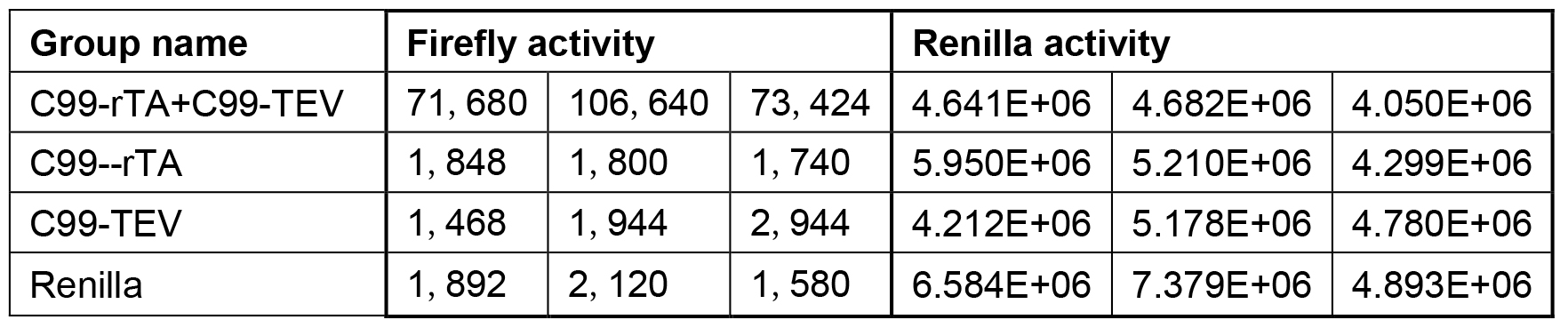
Table 2. Relative activity and Normalized activity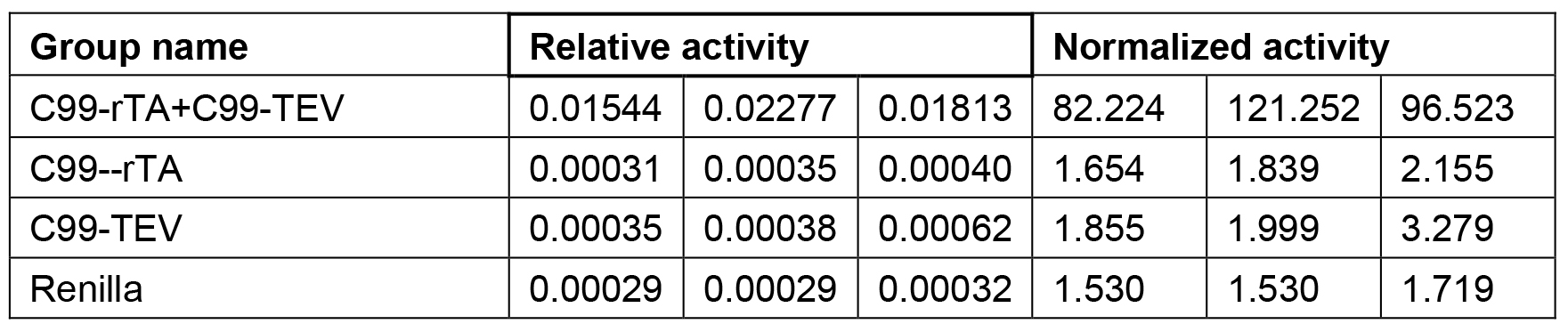
Note: For activity normalization, each relative activity is divided by the average of the relative activity of WT (C99-T4L-rTA) and times 100.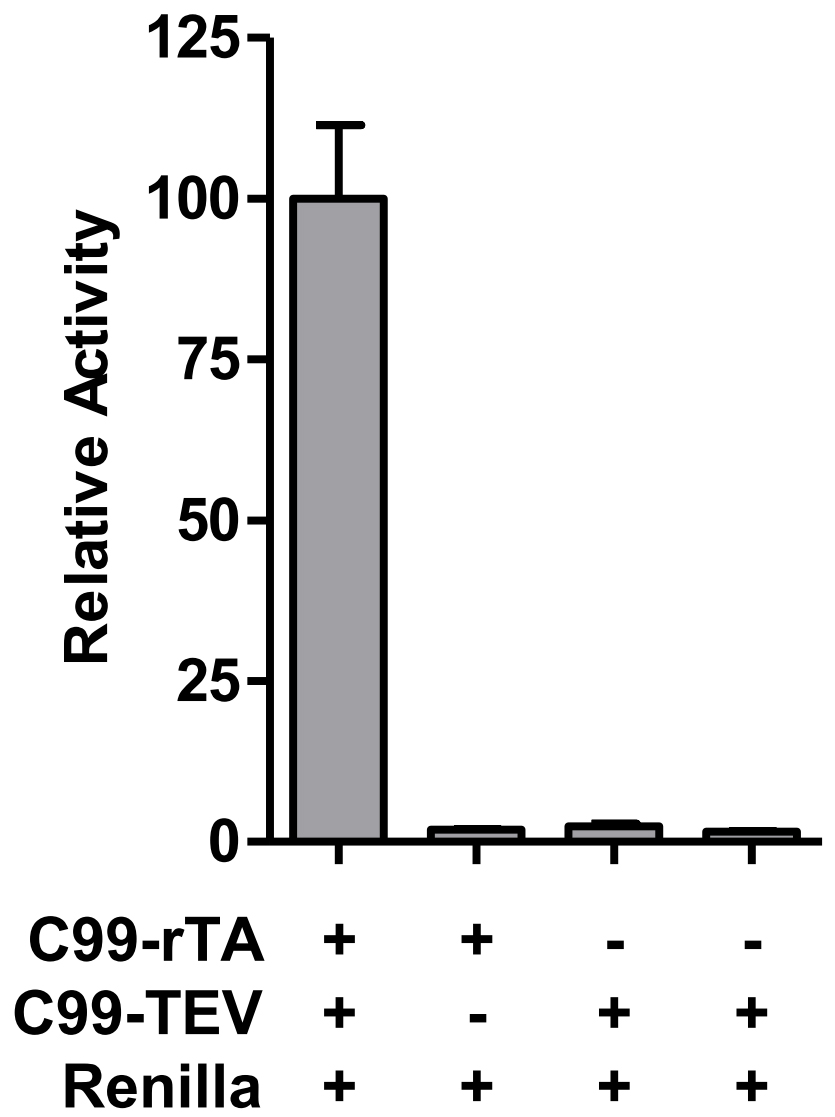
Figure 2. Validation of C99 dimerization by Tango assay
Notes
The status of the cells is very important. When performing data analysis, pay attention to the Renilla luciferase signal (Table 3). It should be around 1,000,000 photo counts.
Table 3. Examples of bad cell status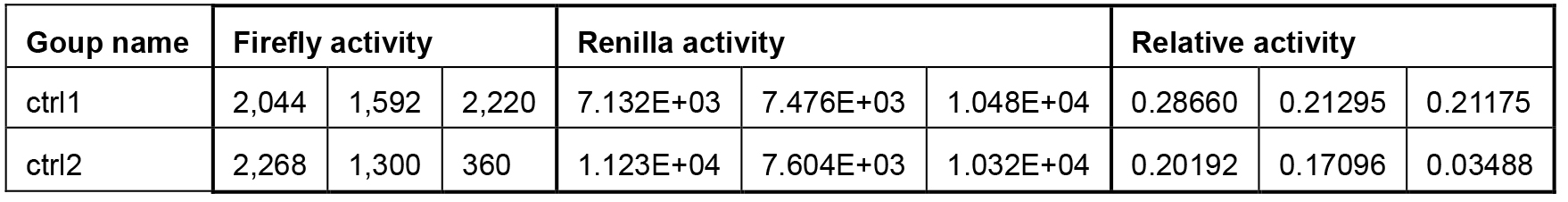
These wells (three wells of Group ctrl1 and three wells of Group ctrl2 should be the negative control, the low Renilla activity, which to some degree reflects the cell status and transfection efficiency, makes the relative activity much higher than normal.
Recipes
- 10x phosphate buffered saline (PBS buffer) (1 L)
11.5 g Na2HPO4
2 g KH2PO4
80 g NaCl
2 g KCl
Dissolve in 1 L of sterile, deionized water
The pH of 1x PBS should be 7.4 - 1x passive lysis buffer (PLB)
Add 1 volume of 5x PLB from Dual-luciferase reporter assay kit to 4 volumes of distilled water and mix well
The 1x PLB can be stored at 4 °C no more than one month - LAR2 substrate (from Dual-luciferase reporter assay kit)
Resuspend the lyophilized Luciferase Assay Substrate in Luciferase Assay Buffer 2 (10 ml for one bottle) and store at -20 °C less than 1 month or -70 °C less than 1 year - Stop & Glo® Reagent (from Dual-luciferase reporter assay kit)
Freshly add 1 volume Stop & Glo® Substrate to 49 volumes Stop & Glo® Buffer and vortex 10 sec
Acknowledgments
This work was supported by the Van Andel Research Institute, the National Natural Science Foundation of China (31300607, 31300245 and 91217311), Ministry of Science and Technology grants 2012ZX09301001, 2012CB910403, and 2013CB910600, XDB08020303, 2013ZX09507001, Shanghai Science and Technology Committee (13ZR1447600), Shanghai Rising-Star Program (14QA1404300), and the National Institute of Health grants DK071662 (H.E.X.), GM102545 and GM104212 (K. M.). The authors declare no conflict of interest.
References
- Barnea, G., Strapps, W., Herrada, G., Berman, Y., Ong, J., Kloss, B., Axel, R. and Lee, K. J. (2008). The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci U S A 105(1): 64-69.
- Xu, T. H., Yan, Y., Kang, Y., Jiang, Y., Melcher, K. and Xu, H. E. (2016). Alzheimer's disease-associated mutations increase amyloid precursor protein resistance to γ-secretase cleavage and the Aβ42/Aβ40 ratio. Cell Discov 2: 16026.
- Yan, Y., Xu, T. H., Harikumar, K. G., Miller, L. J., Melcher, K. and Xu, H. E. (2017). Dimerization of the transmembrane domain of amyloid precursor protein is determined by residues around the gamma-secretase cleavage sites. J Biol Chem 292(38):15826-15837.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yan, Y., Xu, T., Harikumar, K. G., Miller, L. J., Melcher, K. and Xu, H. E. (2017). Detection of Membrane Protein Interactions by Cell-based Tango Assays. Bio-protocol 7(22): e2903. DOI: 10.21769/BioProtoc.2903.
- Yan, Y., Xu, T. H., Harikumar, K. G., Miller, L. J., Melcher, K. and Xu, H. E. (2017a). Dimerization of the transmembrane domain of amyloid precursor protein is determined by residues around the gamma-secretase cleavage sites. J Biol Chem, 292: 15826-15837.
Category
Molecular Biology > Protein > Protein-protein interaction
Biochemistry > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










