- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Characterizing the Transcriptional Effects of Endolysin Treatment on Established Biofilms of Staphylococcus aureus
Published: Vol 8, Iss 12, Jun 20, 2018 DOI: 10.21769/BioProtoc.2891 Views: 6038
Reviewed by: Modesto Redrejo-RodriguezBenoit ChassaingAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Monitoring Protein Splicing Using In-gel Fluorescence Immediately Following SDS-PAGE
Joel Weinberger II and Christopher W. Lennon
Aug 20, 2021 4316 Views

Fluorescent Binding Protein Sensors for Detection and Quantification of Biochemicals, Metabolites, and Natural Products
Salete M. Newton and Phillip E. Klebba
Nov 20, 2022 2906 Views

Framework for Analyzing the Anti-biofilm and Anti-virulence Activities of Fatty Acids from Hermetia illucens Larvae Targeting Multidrug-Resistant Klebsiella pneumoniae
Heakal Mohamed [...] Sergey Leonov
Mar 5, 2026 25 Views
Abstract
Biofilms are the most common lifestyle of bacteria in both natural and human environments. The organized structure of these multicellular communities generally protects bacterial cells from external challenges, thereby enhancing their ability to survive treatment with antibiotics or disinfectants. For this reason, the search for new antibiofilm strategies is an active field of study. In this context, bacteriophages (viruses that infect bacteria) and their derived proteins have been proposed as promising alternatives for eliminating biofilms. For instance, endolysins can degrade peptidoglycan and, ultimately, lyse the target bacterial cells. However, it is important to characterize the responses of bacterial cells exposed to these compounds in order to improve the design of phage-based antimicrobial strategies.
This protocol was developed to examine the transcriptional responses of Staphylococcus aureus biofilm cells exposed to endolysin treatment, as previously described in Fernández et al. (2017). However, it may be subsequently adapted to analyze the response of other microorganisms to different antimicrobials.
Background
It is becoming increasingly clear that subinhibitory doses of antimicrobials may have a regulatory effect on different phenotypes of the target microbes, including biofilm formation, metabolism or virulence. Therefore, studying the potential impact of a novel compound on the target cells at low-level concentrations should be a part of the development process. Indeed, a very effective antibacterial agent that triggers production of virulence factors or antibiotic resistance determinants may not be a good candidate for therapeutic application. On the other hand, considering the physiological differences between biofilm and planktonic cells, it seems logical that the effect of new antibiofilm agents should be analyzed on biofilm-forming cells. Here, we describe a protocol for the analysis of transcriptional responses of biofilm cells upon exposure to subinhibitory concentrations of endolysins, phage-derived proteins that show great promise as biofilm removal agents. Thus, the transcriptome of endolysin-treated cells was compared to control cells by RNA-seq and differential expression of selected genes was later confirmed by RT-qPCR.
Materials and Reagents
- Standard Petri dishes (Labbox, catalog number: PDIP-09N-500 )
- Sterile 10 ml polystyrene culture tubes (Deltalab, catalog number: 300903 )
- Cuvettes for OD600 reading (Deltalab, catalog number: 303103 )
- 1.5 ml microcentrifuge tubes (SARSTEDT, catalog number: 72.690.001 )
- 12-well microtiter plates with Nunclon Delta surface (Thermo Fisher Scientific, Nunc, catalog number: 150628 )
- Sterile plastic loops (1 μl) (VWR, catalog number: 612-9351 )
- MicroAmp® Fast optical 96-well reaction plate with barcode (Thermo Fisher Scientific, Applied Biosystems, catalog number: 4346906 )
- MicroAmp® optical adhesive film (Thermo Fisher Scientific, Applied Biosystems, catalog number: 4311971 )
- Frozen stock of Staphylococcus aureus (for example, S. aureus IPLA1 from our laboratory collection) stored in glycerol at -80 °C
- Filtered LysH5 endolysin stock stored in NaPi buffer with 30% glycerol at -80 °C (~350 μg/ml = 5.8 μM) purified as described previously (Gutiérrez et al., 2014)
- Agarose for electrophoresis (Conda, catalog number: 8008 )
- Glass beads, acid washed (≤ 106 µm, sterile) (Sigma-Aldrich, catalog number: G4649 )
- RNA protect® Bacteria Reagent (QIAGEN, catalog number: 76560 )
- IllustraTM RNAspin Mini Kit (GE Healthcare, catalog number: 25050071 )
- Chloroform (Merck, catalog number: 1024451000 )
- Ethanol (Fisher Scientific, catalog number: BP28184 )
- SUPERase-InTM RNase Inhibitor (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM2694 )
- Turbo DNA-free kitTM (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM1907 )
- DL-Dithiothreitol (Sigma-Aldrich, catalog number: D0632-5G )
- Phenol, Molecular Biology Grade (Merck, Calbiochem, catalog number: 516724-100GM )
- iScriptTM Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories, catalog number: 1708841 )
- Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4367659 )
- Bacteriological agar (ROKO S.A.)
- D(+)-Glucose (Merck, catalog number: 1.08337.1000 )
- Sodium chloride (NaCl) (Merck, catalog number: 1.06404.1000 )
- Potassium chloride (KCl) (VWR, BDH, catalog number: 437025H )
- Sodium phosphate dibasic (Na2HPO4) (VWR, AnalaR NORMAPUR, catalog number: 102495D )
- Potassium phosphate monobasic (KH2PO4) (Merck, catalog number: 1048731000 )
- Sodium dihydrogen phosphate monohydrate (NaH2PO4·H2O) (ITW Reagents Division, AppliChem, catalog number: 131965.1211 )
- UltraPureTM Tris Buffer (Thermo Fisher Scientific, catalog number: 15504020 )
- Glacial acetic acid (Merck, catalog number: 1.00063.2500 )
- 0.5 M EDTA (pH 8.0) (Alfa Aesar, USB, catalog number: J15701 )
- TSB medium (tryptic soy broth, Scharlab, catalog number: 02-200-500 ) (see Recipes)
- TSA agar plates (see Recipes)
- TSB medium supplemented with glucose (TSBG) (see Recipes)
- Phosphate buffered saline (PBS) solution (see Recipes)
- Sodium phosphate (NaPi) buffer (see Recipes)
- Tris-acetate-EDTA (TAE) buffer (see Recipes)
Equipment
- Pipettes (volume ranges: 1 μl-10 μl, 2 μl-20 μl, 20 μl-200 μl, 200 μl-1,000 μl)
- Shaking (250 rpm) and static incubators at 25 °C and 37 °C
- Spectrophotometer
Note: It is used to measure optical density (OD600) of cell culture. - Epoch microplate spectrophotometer (BioTek Instruments, model: Epoch )
- Refrigerated centrifuge (Eppendorf, model: 5415 R )
- FastPrep®-24 (MP Biomedicals, catalog number: 116004500 )
- Gel electrophoresis apparatus (Bio-Rad Laboratories, Mini-Sub® Cell GT Cell)
- Vortex
- 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Applied Biosystems, catalog number: 4351107 )
- Illumina HiSeq2000 platform
- Computer equipped with four Intel Xeon E5-4650 v2 2.4GHz 25M 8GT/s 10-core processors, 256 GB RAM, and running CentOS Linux release 7.3.1611
Note: The computer is for carrying out computation.
Software
- FastQC (http://www.bioinformatics.babraham.ac.uk/projects/download.html#fastqc)
- BowTie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) (Langmead and Salzberg, 2012)
- EDGE-Pro (http://ccb.jhu.edu/software/EDGE-pro/) (Magoc et al., 2013)
- DEseq2 (http://bioconductor.org/packages/release/bioc/html/DESeq2.html) (Love et al., 2014)
Procedure
- Biofilm formation and treatment (Figure 1)
- Streak out S. aureus strain (IPLA1) from the frozen stock onto a TSB agar plate and incubate statically overnight at 37 °C.
- To obtain three biological replicates, pick 3 isolated colonies of S. aureus from the agar plate with a sterile plastic loop and inoculate into three 10-ml polystyrene tubes containing 2 ml of TSB medium.
- Grow bacterial cultures overnight at 37 °C, with shaking at 250 rpm.
- Dilute the overnight cultures to an OD600 of 0.1 in TSBG medium (TSB supplemented with glucose), containing approximately 107 CFU/ml, and then make a 1:20 dilution in the same medium to prepare the inoculum for the biofilm assays.
- Inoculate 2 ml from this cell suspension (approximately 5 x 105 CFU/ml) into each well of a 12-well microtiter plate (four wells per biological replicate).
- Incubate the microtiter plate in static for 24 h at 25 °C.
Note: In this case, the temperature used for biofilm formation and treatment was 25 °C, which represents treatment/disinfection at “room temperature”. Nonetheless, the experiment could have also been performed at different temperatures; for instance, at 37 °C to represent treatment of human infection. - Remove the planktonic phase from the wells and wash the biofilms twice each with 2 ml of PBS.
- For each replicate, add 1 ml of NaPi buffer alone to two wells and 1 ml of NaPi containing 10.94 μg/ml (0.18 μM) of LysH5 to the other two wells.
- Incubate in static for 30 min at 25 °C.
- Remove supernatant.
- Wash twice with PBS.
- Harvest cells corresponding to the same biological replicate and treatment in 1 ml of RNA protect® and 500 μl PBS by scraping with a pipette tip and transfer to a clean Eppendorf tube.
- Process the samples according to the RNA protect® manufacturer’s instructions.
- Store at -80 °C or proceed to RNA purification.
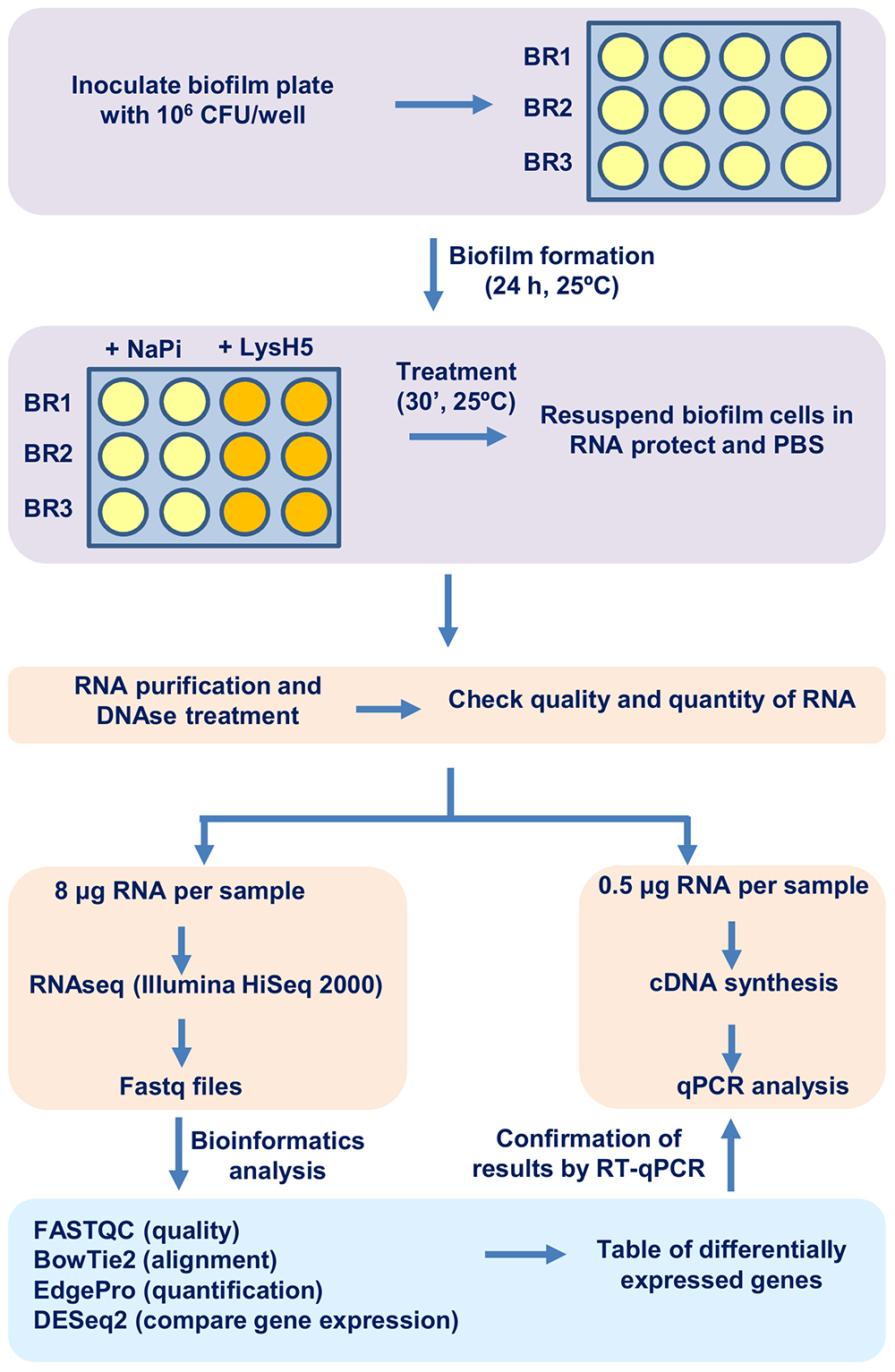
Figure 1. Schematic representation of the protocol. The different steps of this method include biofilm formation and treatment (purple), subsequent RNA purification and RNA-sequencing (pink) and, finally, computer analysis of the generated data (light green).
- Streak out S. aureus strain (IPLA1) from the frozen stock onto a TSB agar plate and incubate statically overnight at 37 °C.
- RNA purification and sequencing (Figure 1)
- To achieve cell lysis, perform mechanical disruption of the cells with glass beads and phenol using FastPrep equipment.
- After lysis, samples were centrifuged at 9,000 x g for 10 min at 4 °C.
- Transfer the upper phase to a clean tube and add 500 μl chloroform and then centrifuge for 5 min at 9,000 x g and 4 °C.
- Transfer the upper phase to a clean tube and mix with 250 μl of ethanol by pipetting.
- Transfer the samples mixed with ethanol to the columns provided with the illustraTM RNAspin Mini kit and perform the rest of RNA purification steps following the instructions provided by the manufacturer.
- Elute in 50 μl of nuclease-free water and add 1 μl of SUPERase-InTM.
- Add 5 μl of Turbo DNase buffer and 1 μl of Turbo DNase per 50 μl sample and incubate for 30 min at 37 °C.
- Add 1 μl of Turbo DNase per sample and incubate for another 30 min at 37 °C.
- Remove DNase from sample with inactivation reagent as indicated by the manufacturer.
- Add 1 μl of SUPERase inhibitor per 50 μl sample.
- Check RNA quality and concentration by agarose gel electrophoresis (1% agarose) in TAE buffer and the Epoch microplate spectrophotometer (Figure 2). RNA concentrations obtained with this protocol usually range between 200 and 700 ng/μl.
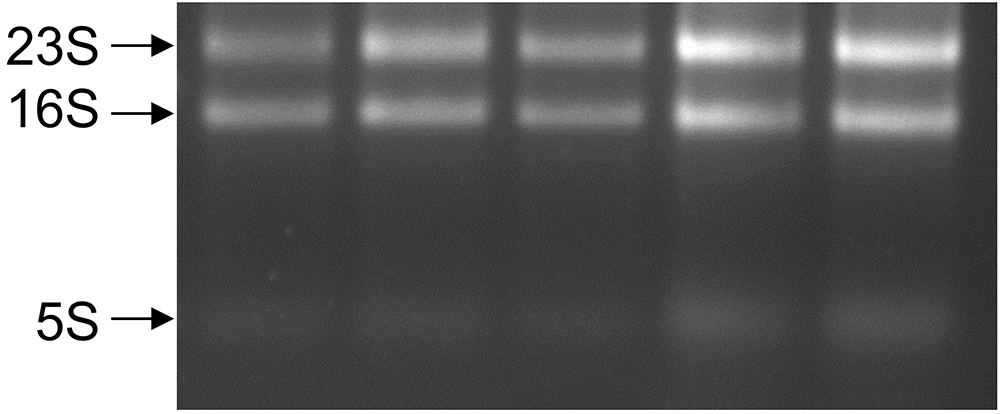
Figure 2. Agarose gel electrophoresis of total RNA from S. aureus biofilm samples. Aliquots (1-2 μl) from different RNA samples were run in a 1% agarose gel. Two bands corresponding to the 23S and 16S rRNAs should be visible and preferably in a proportion of 2:1 (23S:16S) indicating RNA integrity. Sometimes a lower band corresponding to 5S rRNA can also be observed. - Samples with A260/A280 ratios ≥ 1.8 can be considered adequate for RNA-seq analysis. Otherwise, clean up the samples with the illustraTM RNAspin Mini kit following the protocol recommended by the manufacturer.
- Take 8 μg of RNA from each sample and proceed with sequencing steps according to the protocols recommended by the manufacturer of the selected platform. For example, in this study samples were sent to an external service provider (Macrogen Inc., South Korea) for sequencing with an Illumina HiSeq2000 platform according to the protocols recommended by Illumina, generating 100-bp paired-end reads.
- To achieve cell lysis, perform mechanical disruption of the cells with glass beads and phenol using FastPrep equipment.
- Computer analysis of the generated data (Figure 1)
- Check the quality of the reads in FASTQ format with FastQC.
- Download the reference genome in FASTA format (.fa or .fna), the protein table file (.ptt) and the RNA table (.rnt) from the NCBI archive (ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank/Bacteria/).
- Run script “edge.pl” with arguments indicating the FASTQ files containing paired-end reads for each sample (-u and –v) as well as the three files mentioned above (-g, -p and –r) and the prefix for the output files names (-o). In a first step, EDGE-Pro will map the reads to the reference genome using program BowTie2 and create an alignment file as output (this file will be in sequence alignment map or SAM format). BowTie2 also indicates the percentage of alignment to the reference genome. Once completed this step, EDGE-Pro performs transcript quantification into Reads Per Kilobase of transcript per Million mapped reads (RPKMs). The output files containing the RPKM counts will end in “.rpkm_0”.
Example: /edge.pl -g SAreference.fna -p SAreference.ptt -r SAreference.rnt -u Lys_1-1.fastq -o Lys1 -v Lys_1-2.fastq - Run script “edgeToDeseq.perl” indicating the .rpkm_0 files to be analyzed in order to generate a table gathering the raw counts for each gene and each sample. This table will be saved in the output “deseqFile”.
Example: /edgeToDeseq.perl NaPi1.rpkm_0 NaPi2.rpkm_0 NaPi3.rpkm_0 Lys1.rpkm_0 Lys2.rpkm_0 Lys3.rpkm_0
Note: This step is necessary because DESeq2 requires information on raw counts and not RPKMs. - Perform differential expression analysis between treated and untreated samples with DESeq2 by using the “deseqFile” from the previous step as an input. Select genes with an adjusted P-value < 0.05 for further analysis and save the table of differentially-expressed genes in .csv format.
- Check the quality of the reads in FASTQ format with FastQC.
- Confirmation of RNA-seq results by RT-qPCR (Figure 1)
- Convert 0.5 μg RNA from each sample into cDNA with iScriptTM Reverse Transcription Supermix for RT-qPCR as indicated by the manufacturer.
- Dilute cDNA samples 1:25 in nuclease-free water and use them as a template for qPCR.
- To perform qPCR, add 2.5 μl aliquots from the different samples to each well of a MicroAmp® Fast optical 96-well reaction plate together with 3.25 μl of nuclease-free water, 0.25 μl of each primer from a 10 μM stock, and 6.25 μl of Power SYBR® Green PCR Master Mix.
- Analyze each biological replicate in duplicate.
- Determine changes in gene expression by using a reference gene (in this case rplD) according to the 2-ΔΔCT method (Livak and Schmittgen, 2001), in which ΔCT = CT(target gene) - CT(reference gene) and ΔΔCT = ΔCT(target sample) - ΔCT(reference sample).
- Convert 0.5 μg RNA from each sample into cDNA with iScriptTM Reverse Transcription Supermix for RT-qPCR as indicated by the manufacturer.
Data analysis
For reproducibility, it is recommended to analyze three independent biological replicates (BR). Statistical analysis of RNAseq data was performed as part of the differential gene expression analysis with the DESeq2 package, and only those genes with adjusted P-values < 0.05 were selected for further analysis. Regarding fold-change, we normally set the cut-off at 2-fold change (log2 fold-change = 1). However, in this case all genes displaying significant changes based on the adjusted P-values were analyzed further. The small changes are probably due to the fact that only part of the biofilm population was exposed to the antimicrobial. In addition to confirming the genes under the conditions described here, changes were further evaluated in a liquid culture exposed to endolysin LysH5. This analysis showed more evident changes in some of the genes identified by RNA-seq, which reinforced the idea that the transcriptional changes observed in the biofilm were indeed a result of endolysin exposure.
Recipes
- Tryptic soy broth (TSB)
30 g TSB medium
Dissolve in 1 L ddH2O and autoclave - TSA agar plates
TSB medium with 2% agar
Dissolve in ddH2O and autoclave - TSBG medium
TSB medium with 0.25% glucose
Dissolve in ddH2O and autoclave - Phosphate buffered saline (PBS) solution
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
2 mM KH2PO4
Adjust pH to 7.4
Dissolve in ddH2O and autoclave - NaPi buffer
50 mM sodium phosphate
Adjust pH to 7.4
Dissolve in ddH2O and autoclave - TAE buffer (50x stock solution)
242 g of Tris
57.1 ml of glacial acetic acid
100 ml of 0.5 M EDTA (pH 8.0)
Add deionized water to 1 L
Acknowledgments
The development of this protocol was funded by grant AGL2012-40194-C02-01 (Ministry of Science and Innovation, Spain), AGL2015-65673-R (Program of Science, Technology and Innovation 2013-2017), Proyecto Intramural CSIC 201770E016, EU ANIWHA ERA-NET (BLAAT ID: 67), and GRUPIN14-139 (FEDER EU funds, Principado de Asturias, Spain). L.F. was awarded a Marie Curie Clarin-Cofund postdoctoral fellowship. P.G. and A.R. are members of the FWO Vlaanderen-funded PhageBiotics Research community (WO.016.14) and the bacteriophage network FAGOMA. This protocol was adapted from the previously published article Fernández et al. (2017).
Competing interests
The authors declare that they have no conflict of interest.
References
- Fernández, L., González, S., Campelo, A. B., Martínez, B., Rodríguez, A. and García, P. (2017). Downregulation of autolysin-encoding genes by phage-derived lytic proteins inhibits biofilm formation in Staphylococcus aureus. Antimicrob Agents Chemother 61(5).
- Gutiérrez, D., Ruas-Madiedo, P., Martínez, B., Rodríguez, A. and García, P. (2014). Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS One 9(9): e107307.
- Langmead, B. and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4): 357-359.
- Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4): 402-408.
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12): 550.
- Magoc, T., Wood, D. and Salzberg, S. L. (2013). EDGE-pro: Estimated degree of gene expression in prokaryotic genomes. Evol Bioinform Online 9: 127-136.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Fernández, L., González, S., Gutiérrez, D., Campelo, A. B., Martínez, B., Rodríguez, A. and García, P. (2018). Characterizing the Transcriptional Effects of Endolysin Treatment on Established Biofilms of Staphylococcus aureus. Bio-protocol 8(12): e2891. DOI: 10.21769/BioProtoc.2891.
Category
Microbiology > Microbial biofilm > Response to antimicrobials
Molecular Biology > Protein > Activity
Molecular Biology > RNA > RNA sequencing
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









