- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Metals in Whole Cells, Thylakoids and Photosynthetic Protein Complexes in Synechocystis sp. PCC6803
Published: Vol 8, Iss 12, Jun 20, 2018 DOI: 10.21769/BioProtoc.2889 Views: 6331
Reviewed by: Nidhi SharmaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection and Quantification of Heme and Chlorophyll Precursors Using a High Performance Liquid Chromatography (HPLC) System Equipped with Two Fluorescence Detectors
Jan Pilný [...] Roman Sobotka
Feb 5, 2015 13029 Views

Determination of Mn Concentrations in Synechocystis sp. PCC6803 Using ICP-MS
Fabian Brandenburg [...] Marion Eisenhut
Dec 5, 2017 8588 Views

Evaluation of the Condition of Respiration and Photosynthesis by Measuring Chlorophyll Fluorescence in Cyanobacteria
Takako Ogawa and Kintake Sonoike
May 5, 2018 8246 Views
Abstract
Metals are essential in many biological processes, including oxygenic photosynthesis. Here we described a method to measure the metal pool in whole cells and thylakoids, including the bioactive pool in intact photosynthetic protein complexes in the model oxygenic cyanobacterium Synechocystis PCC6803. In the first part of the protocol, whole cells and thylakoid membranes are carefully prepared, in which the total metal concentrations are measured by inductively coupled plasma triple-quadrupole mass spectrometry (ICP-QQQ-MS). In the second part of the protocol, isolated thylakoids are solubilized to release the integral membrane proteins and the metal binding protein complexes. These intact photosynthetic protein complexes are subjected to size exclusion chromatography (SEC) and metal binding in the size separated complexes is analyzed by hyphenation with ICP-QQQ-MS.
Keywords: CyanobacteriaBackground
The process of oxygenic photosynthesis requires metals due to their essential functions as cofactors and catalysts in the photosynthetic electron transport chain. The photosynthetic apparatus requires iron (Fe) in the form of either Fe-S clusters, heme-bridged Fe and non-heme Fe, copper (Cu) in the soluble mobile electron carrier protein plastocyanin, magnesium (Mg) in chlorophylls, calcium (Ca) and manganese (Mn) in the oxygen evolving complex of photosystem II (PSII). Tight control of metal allocation inside photosynthetic cells is essential for cell survival since an imbalanced metal accumulation induces mismetallation and inactivation of the various metallo-enzymes. An accurate analysis of the concentration and allocation of metals in photosynthetic cells is therefore important to investigate the role of key factors and proteins involved in metal homeostasis. While the method described in Brandenburg et al. (2017) accurately quantifies the periplasmic and intracellular pools of Mn in Synechocystis cells, the protocol presented here provides a wider overview, quantifying metals in whole Synechocystis cells, isolated thylakoids, and the bioactive metal pool in fractionated photosynthetic complexes. This protocol is partially based on the method described in Schmidt et al. (2015) for barley thylakoids.
Materials and Reagents
- Pipette tips
- 50 ml conical tubes (Greiner Bio One International, catalog number: 227261 )
- 1.5 and 2 ml Eppendorf tubes (Eppendorf, catalog numbers: 0030120086 and 0030120094 , respectively)
- Microwave Teflon tubes, 8 ml (VWR, catalog number: 525-0178 )
- Nylon membrane filters, 0.45 µm (Frisenette, catalog number: 13NY045-100 )
- Syringe, 1 ml (Frisenette, catalog number: 9161406 )
- NanoVipers fingertight fittings, 150 µm (Thermo Fisher Scientific, catalog number: 6041.5820 )
- Synechocystis sp. PCC6803 GT (glucose tolerant strain, from Dr. Himadri Pakrasi laboratory, Department of Biology, Washington University, St Louis, MO, USA)
- Multi-metal calibration standards for ICP-MS calibration (CPI International, catalog numbers: P/N4400-132565A and P/N4400-132565B )
- EDTA, BioUltra (Sigma-Aldrich, catalog number: E1644-100G )
- Tricine (Sigma-Aldrich, catalog number: T9784-25G )
- Lysozyme (Sigma-Aldrich, catalog number: 62971-50G-F )
- Sucrose, BioExtra (Sigma-Aldrich, catalog number: S7903-1KG )
- Sodium chloride (NaCl), BioExtra (Sigma-Aldrich, catalog number: S7653-250G )
- Magnesium chloride (MgCl2·7H2O), BioExtra (Sigma-Aldrich, catalog number: M2670-1KG )
- Na-ascorbate, BioExtra (Sigma-Aldrich, catalog number: 11140-250G )
- Sodium fluoride (NaF), BioExtra (Sigma-Aldrich, catalog number: S7920-100G )
- Liquid nitrogen
- PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific, catalog number: 23225 )
- Glass beads (Sigma-Aldrich, catalog number: G4649 )
- Milli-Q water
- PlasmaPURE 67-69% HNO3 (SCP SCIENCE, catalog number: 250-039-175 )
- 30% H2O2 (prepare 15% H2O2 with Milli-Q water) (Sigma-Aldrich, catalog number: 31642 )
- n-Dodecyl-β-D-Maltopyranoside (β-DM) (Anatrace, catalog number: D310LA )
- Bis-Tris, BioExtra (Sigma-Aldrich, catalog number: B7535-100G )
- Glycerol, BioExtra (Sigma-Aldrich, catalog number: G6279 )
- Betaine, BioUltra (Sigma-Aldrich, catalog number: 61962-250G )
- Pefabloc SC (Roche Diagnostics, catalog number: 11429868001 )
- Apple leaves, standard reference material (Sigma-Aldrich, catalog number: NIST1515 )
- Glucose (Sigma-Aldrich, catalog number: G8270-1KG )
- Na2S2O3·5H2O (Merck, catalog number: 1.06516.0500 )
- FeNH4 citrate (MP Biomedicals, catalog number: 02158040 )
- Na2CO3 (Sigma-Aldrich, catalog number: S7795-500G )
- K2HPO4 (Sigma-Aldrich, catalog number: P8281 )
- NaNO3 (Sigma-Aldrich, catalog number: S8170-250 )
- MgSO4 (Sigma-Aldrich, catalog number: M2643-500G )
- CaCl2 (Sigma-Aldrich, catalog number: C5670-500G )
- Citric acid (Sigma-Aldrich, catalog number: 251275-100G )
- Na2-EDTA (Sigma-Aldrich, catalog number: E4884-500G )
- H3BO3 (Sigma-Aldrich, catalog number: B6768-500G )
- MnCl2 (Sigma-Aldrich, catalog number: 244589-50G )
- ZnSO4 (Sigma-Aldrich, catalog number: Z0251-100G )
- NaMoO4 (Sigma-Aldrich, catalog number: M1003-100G )
- CuSO4 (Sigma-Aldrich, catalog number: 209198-100G )
- Co(NO3)2 (Alfa Aesar, catalog number: 36418.22 )
- BG11-G (see Recipes)
- 100x BG-FPC (see Recipes)
- 1,000x trace element stock (see Recipes)
- FeNH4 citrate stock (see Recipes)
- Na2CO3 stock (see Recipes)
- K2HPO4 stock (see Recipes)
- EDTA washing buffer (see Recipes)
- Tricine buffer (see Recipes)
- Lysozyme solution (see Recipes)
- Homogenization buffer (see Recipes)
- Tricine-NaF buffer (see Recipes)
- Storage buffer (see Recipes)
- Solubilization buffer (see Recipes)
- Detergent solution (see Recipes)
- Mobile phase buffer (see Recipes)
Equipment
- Pipettes
- Erlenmeyer flasks, 2 L (Fisher Scientific, catalog number: 11961566)
Manufacturer: Pyrex, catalog number: 1130/30D . - Orbital shaker (Eppendorf, New BrunswickTM, model: Innova® 2150, catalog number: M1194-0010 )
- Eppendorf centrifuge 5418 R, refrigerated (Eppendorf, model: 5418 R, catalog number: 5401000013 )
- Eppendorf centrifuge 5430 R, refrigerated (Eppendorf, model: 5430 R, catalog number 5428000210 )
- Beckman Coulter centrifuge (Beckman Coulter, model: Avanti® J-25 ) with type 19 rotor, fixed angle
- 250 ml bottles (Beckman Coulter, catalog number: 325620 )
- Microfluidizer Processor (Microfluidics, model: M-110L )
- Light microscope (ZEISS, model: Axiovert 135 TV )
- Bead mill TissueLyser II (QIAGEN, catalog number: 85300 )
- TissueLyser II adapter set (QIAGEN, catalog number: 69982 )
- Heraeus Fresco 21 Microcentrifuge, refrigerated (Thermo Fisher Scientific, catalog number: 75002425 )
- HPLC Microvials PP, 300 µl and screw cap PP, PFTE (VWR, catalog numbers: 548-0440 and 548-0787 )
- Size exclusion column, Biobasic SEC-1000 analytical column (300 x 7.8 mm) and guard column (30 x 7.8 mm), 5 µm particle size, 1000Å pore size (Thermo Fisher Scientific, catalog number: 73605-307846 and 73605-037821 )
- Pressurized Ultrawave System (Ultrawave system, Milestone Srl, Sorisole, Italy)
- HPLC system, bio-inert (i.e., the mobile phase and samples have no contact with metal parts) (Thermo Fisher Scientific, Dionex, model: UltiMate 3000 )
- DionexTM ICS-5000+ DP Dual Pumps (Thermo Fisher Scientific, model: ICS-5000+ DP )
- ICP-QQQ-MS (Agilent Technologies, model: 8800 Triple Quadrupole ICP-MS )
- Nebulizer, Ari Mist HP (Burgener Research, catalog number: AM HP 5500 )
- Gases for ICP: Liquid Argon, grade 5.0, and reaction gas: 20% Oxygen in Argon (AGA, catalog numbers: 107407 and 714039 )
Software
- MassHunter 4.2 Workstation software 8800 ICP-QQQ data analysis (Agilent Technologies)
Procedure
An overview of the entire protocol is given in Figure 1.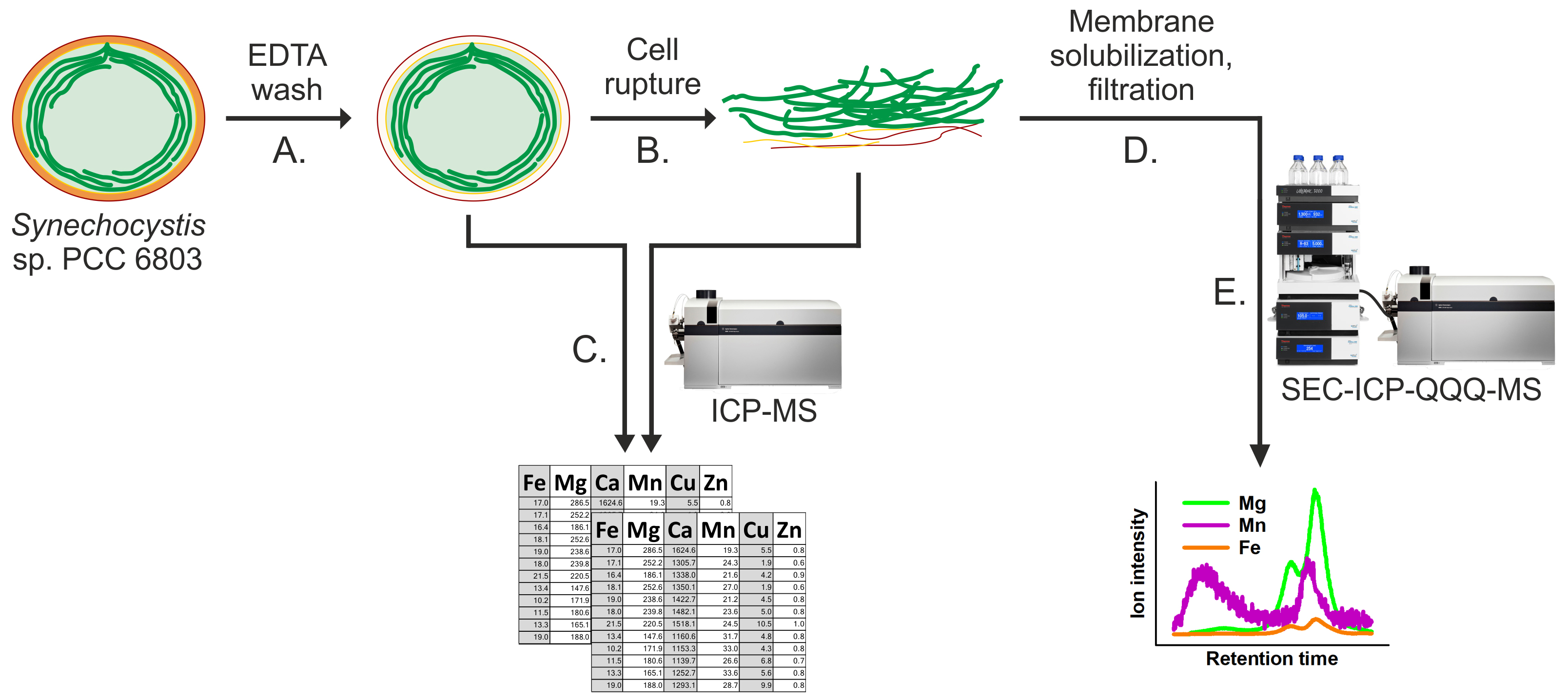
Figure 1. Schematic workflow illustrating the determination of the total metal concentrations in whole cells and thylakoids, and the more detailed analysis of metal allocation into photosynthetic complexes of Synechocystis. Capital letters (A-E) indicate the corresponding procedures in the protocol.
- Whole cell preparation
- Inoculate Synechocystis strains in 0.5 L of BG11-G growth medium (see Recipes), in 2 L Erlenmeyer flasks. Inoculate at least three independent cultures for each strain (biological replicates) to a final 0.05 OD750, following standard microbiological practices.
- Incubate the cultures at a light intensity of 50 µmol photons m-2 sec-1, and shake the flasks on an orbital shaker at 100 rpm until the cultures reach the late exponential phase (0.8-0.9 OD750). Applying these growth conditions the cultures should reach the desired density in about 5 days.
- Before collecting the cultures, measure the OD750 for each culture.
- For each culture, keep aside 50 ml culture for protein concentration determination in whole cells samples (Steps A8-A13).
- Pellet the remaining culture, centrifuging at 5,000 x g for 10 min (at room temperature), using the 250 ml bottles suitable for the type 19 rotor of the Beckman Coulter centrifuge. Repeat the centrifugation step to pellet the rest of the culture volume.
- Wash the cell pellet with 50 ml of EDTA washing buffer (see Recipes), pellet the cells at 5,000 x g for 10 min at room temperature, discard the supernatant and repeat this washing step two more times (Note 1). The supernatant from the first wash will appear slightly colored (with a yellow-orange shade), whereas the supernatant of the third wash should be clear (Figure 2).
- Wash the pellet with 50 ml of Tricine buffer (see Recipes), pellet the cells at 5,000 x g for 10 min, discard the supernatant and repeat this washing step two more times. Before the last centrifugation step, split the cell suspension into two parts, and pellet both cell fractions by centrifugation at 5,000 x g for 10 min at room temperature. One cell pellet constitutes the whole cell preparation ready to be analyzed (see Procedure C). The pellet for the whole cell preparation can be frozen in liquid nitrogen and stored in a freezer until analysis. The second cell pellet is directly further processed to obtain thylakoid membranes (Steps B1-B6).
Note: The metal concentrations will be normalized on protein concentrations. Hence, protein concentration per cell number needs to be determined. For accurate determination of protein concentration in whole cell samples, complete cell rupture is essential. - Treat the cells collected at Step A4 according to Steps A6-A7 and re-suspend the cell pellets to OD750 1 in Tricine buffer (see Recipes).
- Add 1 ml of lysozyme solution (see Recipes) to the cell suspension and gently shake it at room temperature for 30 min. In our hands, this step helps the subsequent rupture.
- Repeatedly rupture the cells in a microfluidizer processor, following manufacturer instruction. Seven to eight cycles are usually sufficient to obtain complete cell rupture.
- Examine the cells rupture under a light microscope to confirm complete rupture of the cells.
- Measure the protein concentration of the samples using the Pierce bicinchoninic acid (BCA) Protein Assay Kit (see Materials and Reagents) for colorimetric detection and quantitation of total protein. The determined protein concentration indicates the µg of proteins in 1 ml of cell lysate, and hence the µg of proteins in 1 ml of culture at 1 OD750.
- Calculate the protein concentration of the whole cell fraction collected in Step A7 by multiplying the protein concentration obtained in Step A12 with the OD750 value measured in Step A3 and by the initial culture volume of the collected cells, which in this case was 225 ml (500 ml of initial culture in Step A1 minus the 50 ml put aside in Step A4, and divided by 2 according to Step A7).
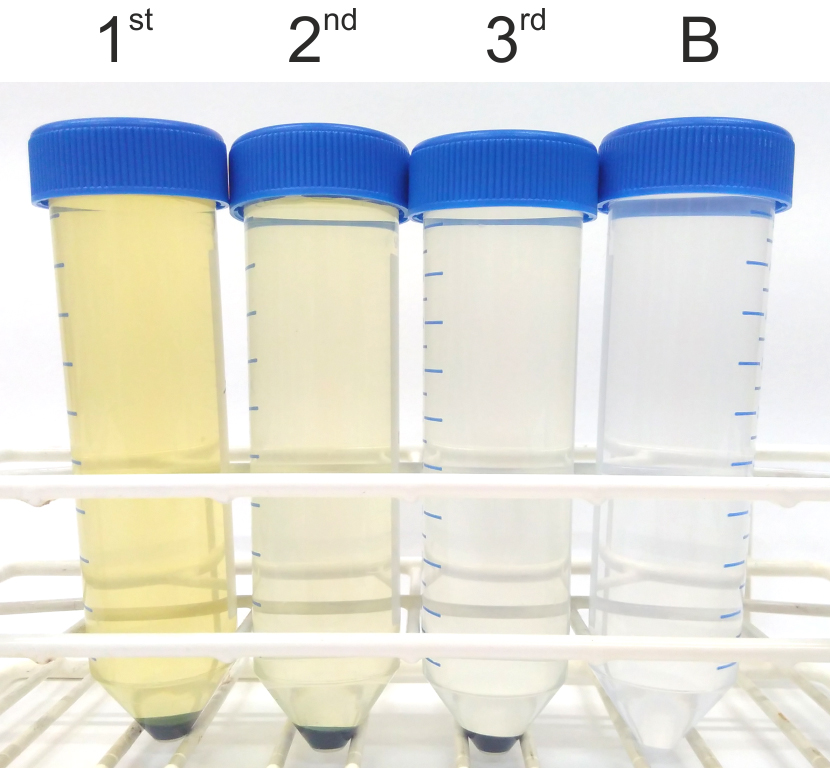
Figure 2. Cell pellet washed three times with EDTA washing buffer. Cell pellets harvested as described in Step A5 are washed three times with the EDTA washing buffer (Step A6). The supernatant of the first wash has a distinct yellow-orange color (1st) which should fade during the second (2nd) and third (3rd) wash. A clear conical tube containing EDTA washing buffer (B) is shown for comparison.
- Inoculate Synechocystis strains in 0.5 L of BG11-G growth medium (see Recipes), in 2 L Erlenmeyer flasks. Inoculate at least three independent cultures for each strain (biological replicates) to a final 0.05 OD750, following standard microbiological practices.
- Thylakoids preparation (Note 2)
Note: In the following steps, samples must be kept on ice, and it is recommended to perform all steps in dim green light to protect the photosynthetic complexes. All centrifugation steps are conducted at 4 °C.- Re-suspend the cell pellets prepared in Steps A1-A7 in 1 ml of homogenization buffer (see Recipes) and transfer the cell suspension to a 2 ml Eppendorf tube holding 1 ml of glass beads.
- Rupture cells in the beads mill, using cooled racks (leave the racks 15 min at -20 °C). The beads mill disrupts the cells through high-speed shaking with glass beads. Cell disruption is performed during six cycles of beads beating, where each cycle involves 1 min of beads beating followed by 1 min of incubation on ice.
- Separate unbroken cells and glass beads by centrifugation for 2 min at 16,000 x g.
- Transfer the supernatant to a 1.5 ml Eppendorf tube and pellet thylakoid membranes for 1 h at 16,000 x g.
- Wash thylakoids two times in 500 µl of Tricine-NaF buffer (centrifuge for 20 min, 16,000 x g) and determine protein concentration using the BCA assay kit.
- Centrifuge thylakoids for 20 min, 16,000 x g and carefully discard the supernatant. The pellet is either digested for determination of total metal analysis (Procedure C) (Note 3) or re-suspended in storage buffer, frozen in liquid N2, and stored at -80 °C for up to 3 months until preparation for SEC-ICP-QQQ-MS analysis (Procedure D).
- Re-suspend the cell pellets prepared in Steps A1-A7 in 1 ml of homogenization buffer (see Recipes) and transfer the cell suspension to a 2 ml Eppendorf tube holding 1 ml of glass beads.
- Determination of total metal concentrations in whole cells and thylakoid membranes
Note: Clean consumables and clean lab procedures must be used in all the following steps. It is recommended to include at least one certified reference (apple [Malus domestica] leaves, NIST 1515; National Institute of Standards and Technology) and one true blank in each digestion run.- Thaw the pellets of whole cells and thylakoid membranes (prepared as described in Procedure A and B, respectively) and add 70% (v/v) HNO3 (see Note 4) directly to each sample in the Eppendorf tubes. Samples are pre-digested for two hours (leave the Eppendorf tubes open, loosely covered by a plastic bag to minimize contamination).
- Carefully transfer (by pipetting) the pre-digested samples to 8 ml Teflon digestion tubes.
- Add 15% (v/v) H2O2 (in a HNO3 to H2O2 ratio of 2.5 to 1, according to Step C1) (see Note 5).
- Digest the samples using a pressurized microwave system (e.g., Milestone, according to the detailed description reported in Hansen et al. [2013]).
- Transfer samples to a clean tube, rinsing the Teflon digestion tubes with Milli-Q water at least three times.
- Dilute samples to a final concentration of 3.5 % HNO3 (v/v) using Milli-Q water (see Note 6). Prepare the calibration standards for the external calibration of the metals of interest (covering a linear range of at least three orders of magnitude and 8 to 10 calibration points).
- Analyze the samples including calibration standards by ICP-MS.
- Thaw the pellets of whole cells and thylakoid membranes (prepared as described in Procedure A and B, respectively) and add 70% (v/v) HNO3 (see Note 4) directly to each sample in the Eppendorf tubes. Samples are pre-digested for two hours (leave the Eppendorf tubes open, loosely covered by a plastic bag to minimize contamination).
- Thylakoid preparation for SEC-ICP-QQQ-MS measurements
Note: In the following steps, samples must be kept on ice, and it is recommended to perform all steps in dim green light to protect the photosynthetic complexes. All centrifugation steps are conducted at 4 °C.- Thaw the thylakoid membrane samples on ice and in darkness.
- Take a homogeneous sub-sample (in this example: corresponding to 300 µg of protein, see Step B5) (pipette gently up and down, avoid vortexing).
- Pellet the thylakoid membranes in a microcentrifuge by centrifugation at 7,000 x g for 2 min.
- Discard the supernatant and re-suspend (by gently pipetting up and down) the thylakoid membranes in 150 µl ice-cold solubilization buffer (freshly prepared, see Recipes) to a final protein concentration of 2 µg/µl.
- Add an equal volume of detergent solution (150 µl, see Recipes) to a final protein concentration of 1 µg/µl and solubilize the thylakoid membranes on ice and in darkness for 10 min.
- Solubilized proteins (supernatant) are separated from insoluble material (pellet) by centrifugation at 16,000 x g for 10 min.
- The solubilized proteins (supernatant) are immediately collected, and passed through a 0.45 µm nylon membrane filter using 1 ml syringes, into HPLC microvials.
- Keep samples on ice and in darkness until the subsequent speciation analysis (Procedure E).
- Thaw the thylakoid membrane samples on ice and in darkness.
- SEC-ICP-QQQ-MS measurements: Determination of metal binding in photosynthetic complexes (speciation analysis)
Note: During speciation analysis, the HPLC autosampler and column compartment are kept cold at 6 °C. This part of the protocol is based on the method developed by Schmidt et al. (2015)- Equilibrate the HPLC system and the size-exclusion column with the mobile phase buffer (see Recipes) for at least 1 h.
- Inject 50 µg of freshly solubilized thylakoid proteins onto the size-exclusion column, using the HPLC system.
- Perform isocratic protein fractionation and elution using mobile phase buffer at a flow rate of 1 ml/min for 35 min.
- For the online detection of metals bound in photosynthetic protein complexes, the column outlet is directly coupled to the ICP-QQQ-MS (Note 7).
- Time-resolved chromatograms with multi-element detection are recorded (Figure 3).
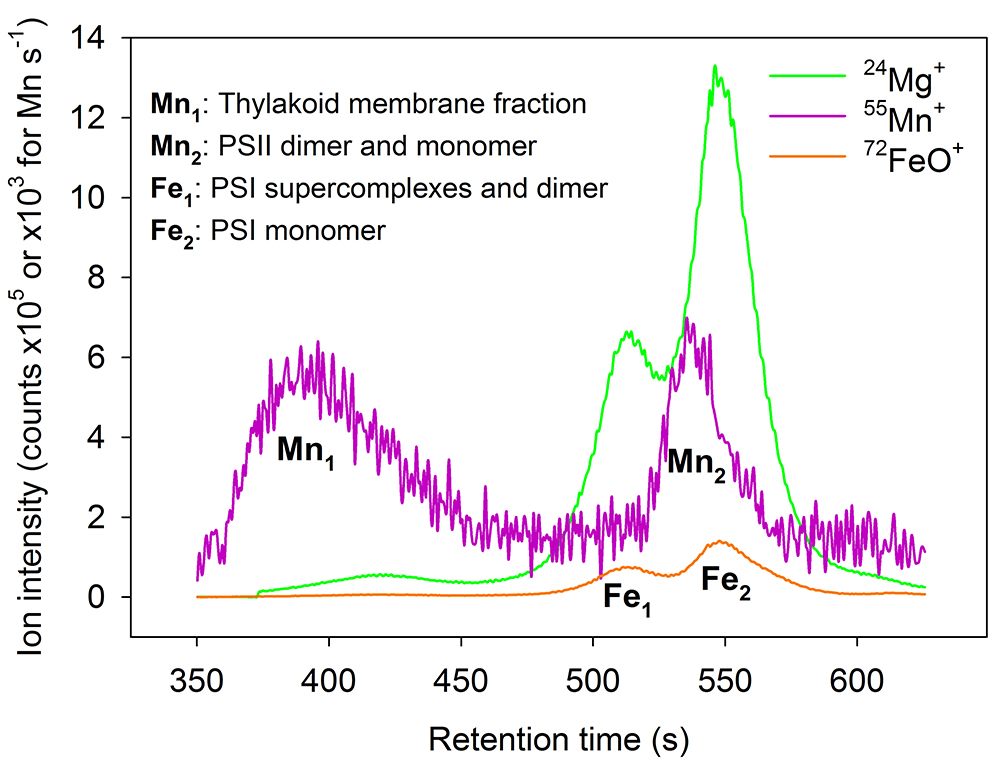
Figure 3. Multi-metal chromatogram. Size-exclusion profiles recorded for the non-oxide ions 24Mg+ and 55Mn+ and the oxide ion 72FeO+. Individual Mn and Fe subfractions are assigned according to Gandini et al. (2017), the Mg profile is illustrated as a proxy for chlorophyll.
- Equilibrate the HPLC system and the size-exclusion column with the mobile phase buffer (see Recipes) for at least 1 h.
Data analysis
- Total metal concentrations
The metal concentrations in whole cells and thylakoid membranes are determined from the external calibration curves for each metal, using the Masshunter software (the software enables automatic calculation of sample concentrations). The data are exported and the metal concentrations are normalized to protein concentrations for each sample. It is recommended to perform two-way analysis of variance (ANOVA) on metal concentrations from at least two independent experiments, each consisting of three independent preparations for each strain of interest, to take into account any variation between experiments. In this respect, we observed that the metal count of a strain differed more between two independent experiments than within independent cultures of the same experiment. This variability could be due to various reasons, such as slightly different log-phases at culture collection. - Speciation analysis
At least three independent samples for each strain are analyzed by SEC-ICP-MS. As for the total metal concentrations analysis, we strongly recommend analyzing at least two independent experiments. SEC-ICP-MS chromatograms can be individually prepared for each metal of interest, or by overlaying more metals to construct a single multi-metal chromatogram. Prepare the chromatogram for one representative sample. Data are presented as ion counts per second as a function of the retention time. The metal profiles may be background corrected or shown as raw data (Figure 3). Individual peaks are assigned according to Gandini et al. (2017).
Notes
- EDTA is used to remove the Mn storage pool located in the outer membrane and periplasmic space (Keren et al., 2002).
- The thylakoid preparation described here is actually composed by outer membrane, plasma membrane and thylakoids. However, thylakoids are, by far, the predominant membrane in the sample. The abundance of thylakoids in the preparation is particularly evident from the obtained green pellet and when the membrane preparation is loaded onto a sucrose step gradient (as described in Omata and Murata, 1985, see Figure 4).
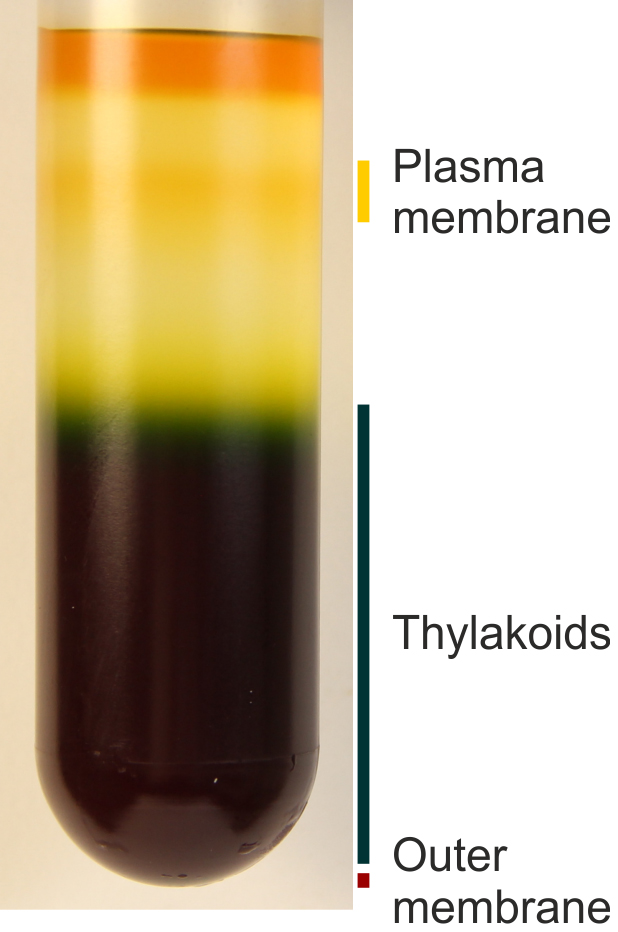
Figure 4. Membrane composition of the thylakoids preparation described in Procedure B. Thylakoids are the most abundant type of membrane, but other membranes are included in the preparation as well (plasma membrane and outer membrane). The sucrose step gradient is prepared according to Omata and Murata, 1985. - If preferred, thylakoid pellets can be frozen in liquid N2 (to ensure the pellet is kept in the bottom of the Eppendorf tube) and stored in a freezer until digestion for total metal analysis.
- In the current protocol, 625 or 50 µl of 70% (v/v) HNO3 was added to the whole cell sample (25 ml OD7501 Synechocystis cells, approximately corresponding to 73 mg of proteins) and thylakoid pellets (corresponding to 0.73 mg protein), respectively. It is important to note, that the given protein amounts may be adjusted (and most likely further reduced). However, make sure that the whole cell and thylakoid pellets are completely covered with HNO3 during pre-digestion.
- Addition of H2O2 facilitates complete digestion at lower temperatures.
- To ensure accurate dilution, the dilution of each sample was performed using an analytical balance. The weight of each sample after dilution (the dilution factor) was noted for each sample for subsequent calculations and data analysis.
- The ICP-QQQ-MS is operated in MS/MS scan mode with oxygen as the reaction gas, enabling the sensitive simultaneous analysis of the oxide and non-oxide ions: 24Mg+, 55Mn+, and 72FeO+. The integration time is set to 0.1 sec per metal. It is recommended to tune the ICP-QQQ-MS for plasma and ion lenses conditions on a daily basis to ensure plasma robustness and maximum sensitivity.
- Synechocystis glucose tolerant strains (see Materials and Reagents) were cultivated in BG11 growth medium that contains glucose (BG11-G, see Recipes) in order to facilitate the study of photosynthetic mutants (Gandini et al. 2017). However, other Synechocystis strains in combination with the BG11 medium but without glucose (Rippka et al. 1979) can be used as well.
Recipes
- BG11-G liquid growth medium, 1 L (see Note 8)
5 ml 1 M Glucose
4.7 g/L Na2S2O3·5H2O
10 ml 100x BG-FPC
1 ml FeNH4 citrate stock
1 ml Na2CO3 stock
1 ml K2HPO4 stock
Bring to 1 L with ultrapure water and sterilize by autoclave - 100x BG-FPC
1.76 M NaNO3
30.4 mM MgSO4
24.5 mM CaCl2
3.12 mM Citric acid
0.279 mM Na2-EDTA
1/10 (v/v) 1,000x trace element stock - 1,000x trace element stock
46.3 mM H3BO3
9.15 mM MnCl2
0.77 mM ZnSO4
1.61 mM NaMoO4
0.32 mM CuSO4
0.17 mM Co(NO3)2 - FeNH4 citrate stock
6 mg/ml FeNH4 citrate
Sterilize by autoclave - Na2CO3 stock
198 mM Na2CO3
Sterilize by autoclave - K2HPO4 stock
175 mM K2HPO4
Sterilize by autoclave - EDTA washing buffer
5 mM EDTA
20 mM Tricine, pH 7.9 - Tricine buffer
20 mM Tricine, pH 7.9 - Lysozyme solution
10 mg/ml Lysozyme - Homogenization buffer
0.4 M sucrose
10 mM NaCl
5 mM MgCl2
20 mM Tricine, pH 7.9
Freshly add: 10 mM Na-ascorbate, 10 mM NaF - Tricine-NaF buffer
5 mM Tricine, pH 7.9
Freshly add: 10 mM NaF - Storage buffer
0.4 M sucrose
10 mM NaCl
5 mM MgCl2
20 mM Tricine, pH 7.9
20% glycerol
Freshly add: 10 mM NaF - Solubilization buffer
25 mM Bis-Tris (pH 7/HCl)
12.5% (v/v) glycerol
2 M Betaine
0.25 mg/ml pefabloc SC - Detergent solution
Note: The detergent solution is prepared from a 10% β-DM stock stored at -20 °C.
2% (w/v) β-DM prepared in solubilization buffer - Mobile phase buffer
25 mM Bis-Tris (pH 7/TFA)
0.03% β-DM
Acknowledgments
This research was supported by Independent Research Fund Denmark – Technology and Production Sciences (grant No. DFF-5054-00042 to S.B.S.).
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Brandenburg, F., Schoffman, H., Keren, N. and Eisenhut, M. (2017). Determination of Mn concentration in Synechocystis sp. PCC6803 using ICP-MS. Bio-protocol 7(23): e2623.
- Gandini, C., Schmidt, S. B., Husted, S., Schneider, A. and Leister, D. (2017). The transporter SynPAM71 is located in the plasma membrane and thylakoids, and mediates manganese tolerance in Synechocystis PCC6803. New Phytol 215(1): 256-268.
- Hansen, T. H., de Bang, T. C., Laursen, K. H., Pedas, P., Husted, S. and Schjoerring, J. K. (2013). Multielement plant tissue analysis using ICP spectrometry. Methods Mol Biol 953: 121-141.
- Keren, N., Kidd, M. J., Penner-Hahn, J. E. and Pakrasi, H. B. (2002). A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry 41(50): 15085-15092.
- Omata, T. and Murata, N. (1985). Electron-transport reactions in cytoplasmic and thylakoid membranes prepared from the cyanobacteria (blue-green algae) Anacystis nidulans and Synechocystis PCC 6714. Biochim Biophys Acta 810(3): 354-361.
- Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M. and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111(1): 1-61.
- Schmidt, S. B., Persson, D. P., Powikrowska, M., Frydenvang, J., Schjoerring, J. K., Jensen, P. E. and Husted, S. (2015). Metal binding in photosystem II super- and subcomplexes from barley thylakoids. Plant Physiol 168(4): 1490-1502.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gandini, C., Husted, S. and Schmidt, S. B. (2018). Analysis of Metals in Whole Cells, Thylakoids and Photosynthetic Protein Complexes in Synechocystis sp. PCC6803. Bio-protocol 8(12): e2889. DOI: 10.21769/BioProtoc.2889.
Category
Plant Science > Plant biochemistry > Other compound
Plant Science > Plant physiology > Photosynthesis
Biochemistry > Other compound > Elements
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










