- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Intracellular and Mitochondrial Reactive Oxygen Species Measurement in Primary Cultured Neurons
Published: Vol 8, Iss 11, Jun 5, 2018 DOI: 10.21769/BioProtoc.2871 Views: 11858
Reviewed by: Xi FengWelsch Charles JeremyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2900 Views

Two-photon (2P) Microscopy to Study Ca2+ Signaling in Astrocytes From Acute Brain Slices
Annamaria Lia and Micaela Zonta
Jul 5, 2025 2875 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 483 Views
Abstract
Reactive oxygen species (ROS) are chemically reactive oxygen containing molecules. ROS consist of radical oxygen species including superoxide anion (O2•−) and hydroxyl radical (•OH) and non-radical oxygen species such as hydrogen peroxide (H2O2), singlet oxygen (O2). ROS are generated by mitochondrial oxidative phosphorylation, environmental stresses including UV or heat exposure, and cellular responses to xenobiotics (Ray et al., 2012). Excessive ROS production over cellular antioxidant capacity induces oxidative stress which results in harmful effects such as cell and tissue damage. Sufficient evidence suggests that oxidative stresses are involved in cancers, cardiovascular disease, and neurodegenerative diseases including Alzheimer’s disease and Parkinson disease (Waris and Ahsan, 2006). Though excessive level of ROS triggers detrimental effects, ROS also have been implicated to regulate cellular processes. Since ROS function is context dependent, measurement of ROS level is important to understand cellular processes (Finkel, 2011). This protocol describes how to detect intracellular and mitochondrial ROS in live cells using popular chemical fluorescent dyes.
Keywords: Reactive oxygen species (ROS)Background
ROS are important to maintain homeostasis in our bodies (Brieger et al., 2012). Many diseases such as cancer, neurodegenerative disease, cardiovascular disease, and diabetics are associated with ROS (Datta et al., 2000). DNA damage caused by ROS is a major cause of accelerating carcinogenesis process, and therapeutic agents targeting ROS have been actively developed (Trachootham et al., 2009). In circulatory system, abnormal oxidative stress increases the production of ROS, leading to various cardiovascular diseases (Forstermann, 2008). Signaling related to diabetes is sensitive to ROS, and these signaling abnormalities induced by abnormal levels ROS cause diabetes complications (Baek et al., 2017). Controlling the ROS levels in the brain is one of the most important activities because abnormal levels of ROS can cause diverse brain diseases. Amyloid beta, known as an important factor in Alzheimer’s disease, causes excessive ROS generation in the brain, neuronal damage (Singh et al., 2011), and eventually dementia (Polidori, 2004). Activated microglia produced by ROS which secretes a variety of cytokines result in neuronal death (Heneka et al., 2014).
ROS are generated by small part of oxygen consumed in mitochondria. A principal species of ROS produced in mitochondria is superoxide anion and it is the byproduct of the electron transport chain (Batandier et al., 2002). In order to detect superoxide in mitochondria, MitoSOX red, a mitochondria superoxide indicator, is used. Due to the positive charge on triphenylphosphonium group, MitoSOX red can effectively penetrate phospholipid bilayer, and accumulate into the matrix of mitochondria. Furthermore, hydroethidine of MitoSOX red allows researchers to discriminate the fluorescent signal generated by superoxide-mediated oxidative products from other non-specific signals (Robinson et al., 2006; Baek et al., 2017).
CM-H2DCFDA is a chloromethyl derivative of H2DCFDA (2',7'-dichlorodihydrofluorescein diacetate), a fluorogenic dye that measures hydroxyl, peroxyl and other ROS activity within the cell and can be used to detect the intracellular formation of ROS (Kirkland et al., 2007). Once the fluorescent probe of CM-H2DCFDA permeates cell membrane, intracellular esterases hydrolyze its acetyl groups and it can be retained in the cell. CM-H2DCFDA is more sensitive to oxidation by H2O2 than superoxide (O2•−) (Fowler et al., 2017). CM-H2DCFDA is widely used in physiological and pathophysiological studies including virus infection (Nykky et al., 2014), cancer (Khatri et al., 2015; Liu et al., 2017), and neurodegenerative diseases (Ng et al., 2014). Using CM-H2DCFDA, we can detect intracellular ROS level by flow cytometry/fluorescence measurement and the localization of ROS producing organelle with confocal microscopy (Forkink et al., 2010).
Materials and Reagents
- Glass bottom cell culture dish type 35 mm and dimension 20 mm (Nest Scientific, catalog number: 801001 )
- Cover glasses thickness No. 1 circular size 18 mm Ø (MARIENFELD, catalog number: 0111580 )
- Petri dish, 100 mm Polysterene aseptic non-tissue culture treated (SPL Life Sciences, catalog number: 10095 )
- 15 ml conical tube (SPL Life Sciences, catalog number: 50015 )
- 10 ml Serological pipettes (SPL Life Sciences, catalog number: 91010 )
- 50 ml conical tube (SPL Life Sciences, catalog number: 50050 )
- Cell strainer 70 μm (Corning, Falcon®, catalog number: 352350 )
- Pregnant female Sprague Dawley rats (E17-E18 days gestation, Orient Korea)
- Poly-D-lysine hydrobromide (Sigma-Aldrich, catalog number: P6407-5mg )
- Phosphate buffered saline powder, pH 7.4, for preparing 1 L solutions (Sigma-Aldrich, catalog number: P3813 )
- CM-H2DCFDA (Thermo Fisher Scientific, InvitrogenTM, catalog number: C6827 )
- Dimethyl Sulfoxide(DMSO) (Merck, catalog number: 317275 )
- MitoSOXTM Red Mitochondrial Superoxide Indicator, for live-cell imaging (Thermo Fisher Scientific, InvitrogenTM, catalog number: M36008 )
- Phosphate buffered saline (PBS) powder, pH 7.4, for preparing 1 L solutions, suitable for cell culture(Sigma-Aldrich, catalog number: P3813 )
- Trypsin (2.5%), no phenol red (Thermo Fisher Scientific, GibcoTM, catalog number: 15090046 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10082147 )
- Neurobasal Medium® (Thermo Fisher Scientific, GibcoTM, catalog number: 21103049 )
- B-27TM Supplement (50x), serum free (Thermo Fisher Scientific, GibcoTM, catalog number: 17504044 )
- DMEM High Glucose (4.5 g/L), with L-Glutamine, with Sodium Pyruvate (Capricorn Scientific, catalog number: DMEM-HPA )
- Penicillin/Streptomycin (100x) (PS) (Capricorn Scientific, catalog number: PS-B )
- Amyloid beta peptide 1-42 Human (ANYGEN, catalog number: AGP-8338 )
- CM-H2DCFDA solution (see Recipes)
- MitoSOXTM Red solution (see Recipes)
- Poly-D-lysine hydrobomide solution (see Recipes)
- Prep medium (see Recipes)
- Culture medium (see Recipes)
- Maintain culture medium (see Recipes)
Equipment
- Haemacytometers (MARIENFELD, catalog number: 0630010 )
- Original Portable Pipet-Aid® Pipette Controller (Drummond Scientific, catalog number: 4-000-100 )
- Dressing Scissors (Surgimax Instruments, catalog number: 85-112-12 ) (Figure 1A 1)
- Dissecting Scissors (Surgimax Instruments, catalog number: 85-127-10 ) (Figure 1A 2)
- Dissecting Scissors (Surgimax Instruments, catalog number: 63-175-11 ) (Figure 1A 3)
- Spring Dressing Forceps Sharp (Surgimax Instruments, catalog number: 85-076-11 ) (Figure 1A 4)
- Spring Dressing Forceps Blunt (Surgimax Instruments, catalog number: 85-073-15 ) (Figure 1A 5)
- Multi Purpose Forceps Pointed (Surgimax Instruments, catalog number: 05-177-11 ) (Figure 1A 6)
- Clean bench (HANBAEK Scientific Technology, catalog number: HB-402 )
- Cell culture CO2 incubator (ARA, catalog number: APR150 )
- Water-bath (Grant Instruments, JB Academy, catalog number: JBA18 )
- Centrifuge (Hanil Scientific, catalog number: Combi 514R )
- Confocal microscope with live cell imaging system (Carl Zeiss, model: LSM700 ) (Figures 1B and 1C)
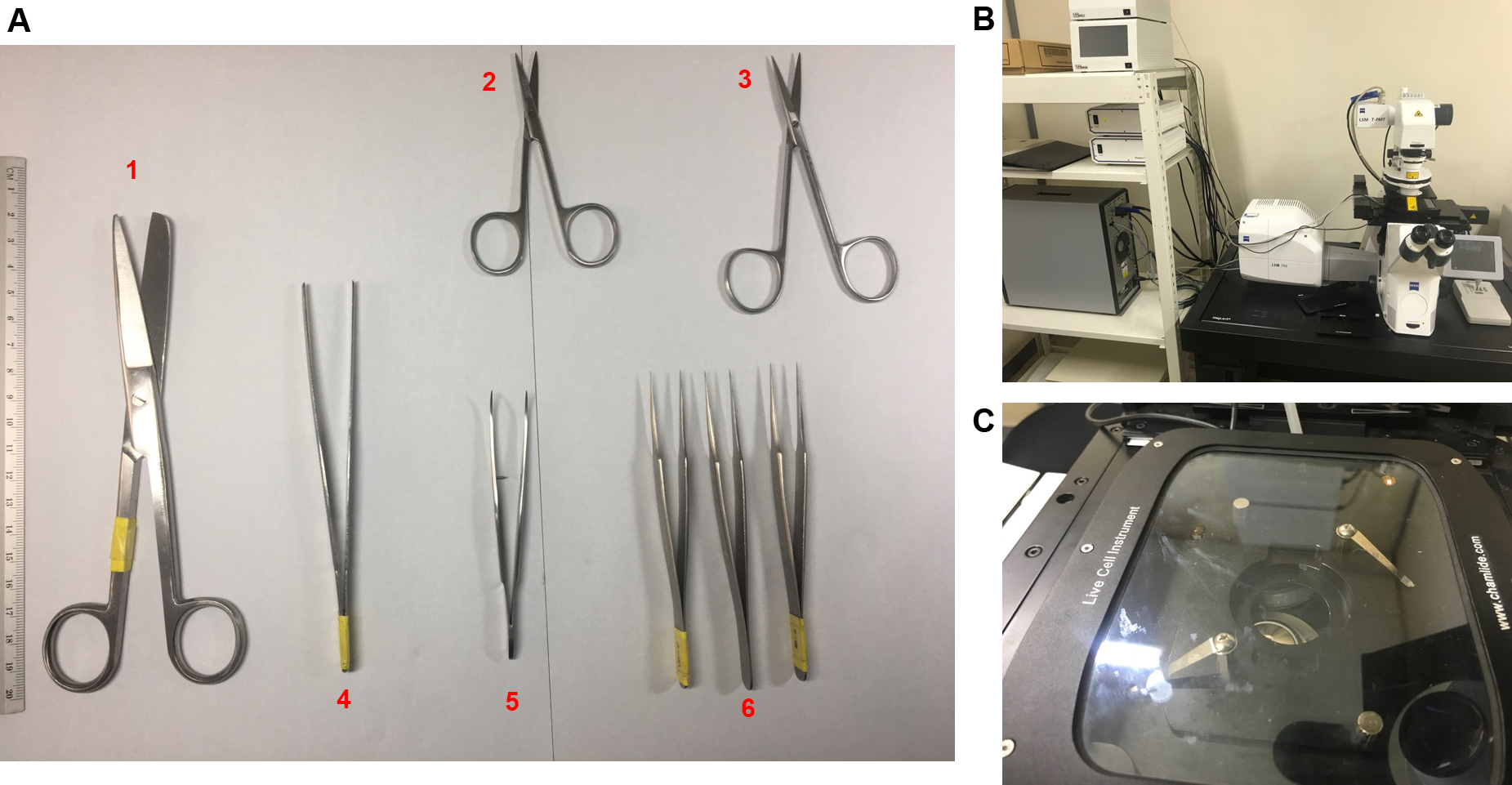
Figure 1. Equipment for the experiment. A. Surgery instruments; B. Confocal microscope (LSM700) with live cell imaging system; C. Live cell chamber.
Software
- For measure
- ZEN black version (ZEISS confocal microscope LSM700 software)
Note: This is default program provided with ZEISS confocal microscope.
- ZEN black version (ZEISS confocal microscope LSM700 software)
- For analysis
- ZEN Blue edition (ZEISS confocal microscope LSM700 software; SR-DIP software for ZEN blue edition)
- ImageJ (ImageJ is an open source image processing program)
- ZEN Blue edition (ZEISS confocal microscope LSM700 software; SR-DIP software for ZEN blue edition)
Procedure
In this protocol, we introduce two types of ROS measurement in primary neuronal cells under oxidative stress.
- In advance, a 35 mm plate for live cell imaging, is coated with 2 ml poly-D-lysine solution at 4 °C for 24 h.
Optional: Instead of using 35 mm dish for ROS measurement, the following method is possible. Place the sterile cover glasses slip flat into each well of a 6-well plate. Add 2 ml of Poly-D-lysine solution to each well and coat at 4 °C for 24 h. - Remove the poly-D-lysine solution and wash twice with cold PBS.
- Dry the coated plate on a bench while E17 rat embryo is being prepared.
- Carefully take out the E17~E18 embryos from the Sprague Dawley rat using Dressing Scissors (Equipment 3) and Spring Dressing Forceps Blunt (Equipment 7). Place them in a Petri dish filled with cold Prep medium (Figures 2A-2D).
Note: In this experiment, the pregnant Sprague Dawley rats were anesthetized with CO2 and euthanized using CO2 after embryos extraction. All experimental procedures were conducted after approval of Institutional Animal Care and Use Committee of Sungkyunkwan University. - Using Dissecting Scissors (Equipment 5) and Spring Dressing Forceps Sharp (Equipment 6), carefully take out individual embryo from uterus and embryonic sack (Figures 2E-2F).
- Using Dissecting Scissors (Equipment 4) and Multi Purpose Forceps Pointed (Equipment 8), excise the scalp and skull of the E17-E18 embryos and pull out the whole brain. Place the extracted brain in Prep medium (Figures 2G-2I).
Note: For proper cell conditions, this process (Steps 4-6) should be finished within 15-20 min.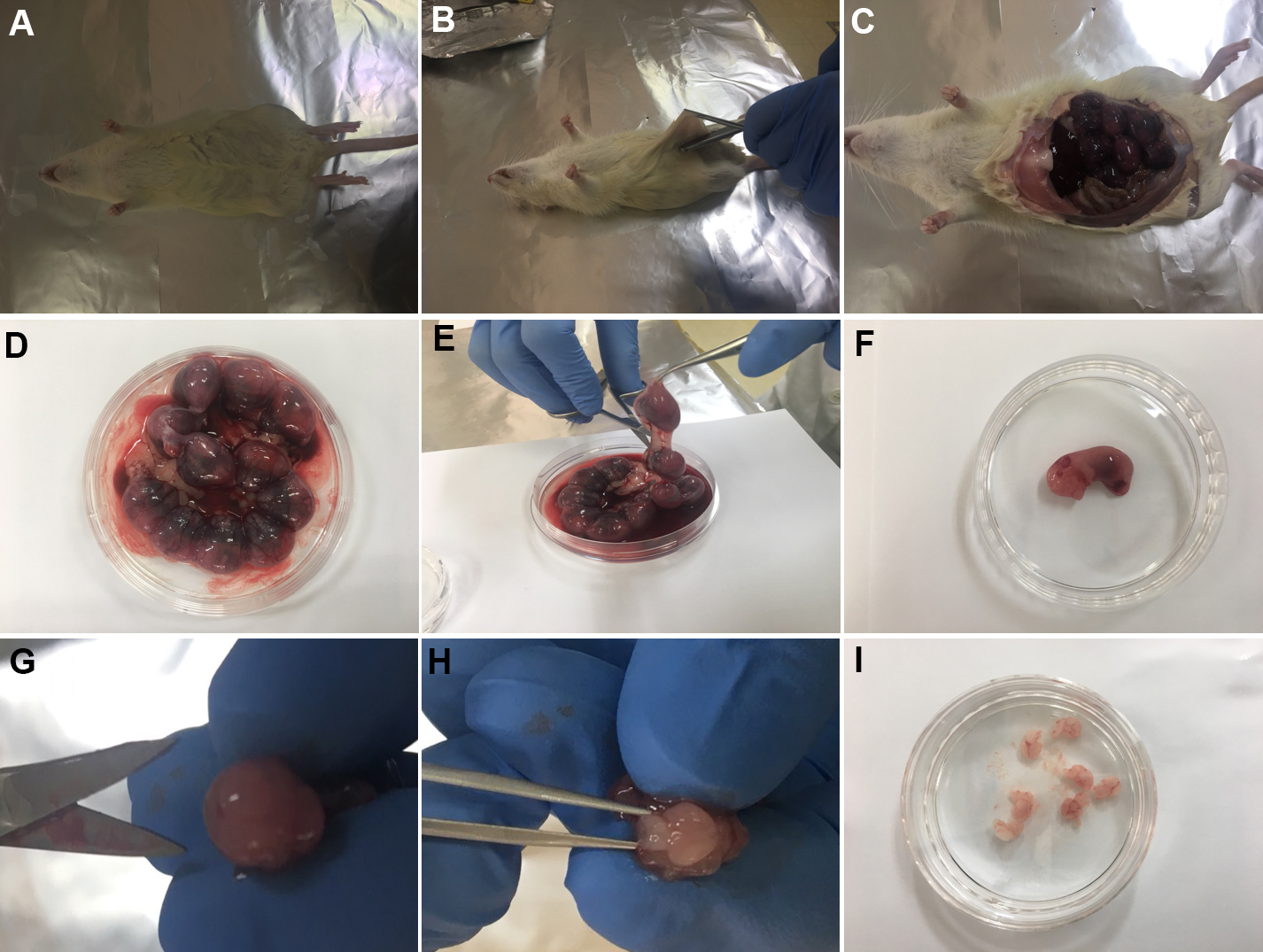
Figure 2. The process of embryo extraction in pregnant Sprague Dawley rat (E17-E18) (Steps 4-6) - After taking out the embryo's brain, place it in the prep medium as shown in Figure 3A.
- Dissect the cerebral cortex from the whole brain as Figure 3B.
Note: When separating the cortex from the embryo’s brain, you must be careful not to separate other parts together. When you progress from Step 7 to Step 8 (Figure 3A to Figure 3B), insert the micro forceps between the inner side of the cortex and the outer part of the striatum, and then cut cortex from embryo’s brain. As you see in Figure 3A, the cortex is on the surface as it envelops other parts of the brain. There is a striatum on the inner side just below the cortex. - Remove meninges and blood vessels outside the cerebral cortex (Figure 3C). Place the cerebral cortex in Prep medium as Figure 3D.
Notes:- After removing the meninges and blood vessels as shown in Figure 3, remove the hippocampus part that is attached to the inner side of the cortex. You can use the cortex part for cortical neuron cell culture or you can conduct hippocampal neuron cell culture with the hippocampus part.
- For proper cell conditions, this process (Steps 7-9) should be done within 20-25 min.
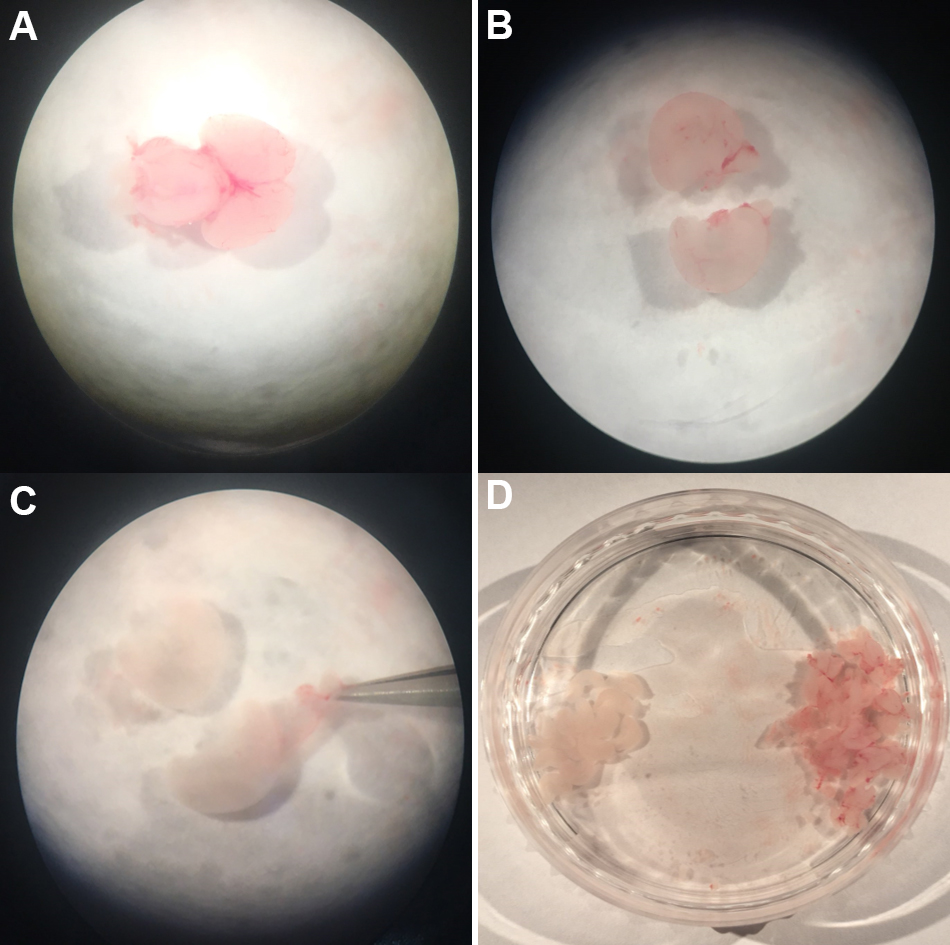
Figure 3. The process of extracting cortex region from the extracted whole brain (Steps 7-9) - After removing the meninges and blood vessels as shown in Figure 3, remove the hippocampus part that is attached to the inner side of the cortex. You can use the cortex part for cortical neuron cell culture or you can conduct hippocampal neuron cell culture with the hippocampus part.
- Transfer the cerebral cortex to a conical tube (15 ml size) filled with 2 ml of Prep medium and add 300 μl of trypsin solution to make final concentration about 10% (Figure 4A).
- Place the conical tube in a 37 °C incubator or water bath and gently tapping it periodically (Figure 4B).
- After about 15 min, carefully suspend the cells using a 10 ml disposable pipette (Figure 4C).
- Put 400 μl of FBS in the tubes to 10% concentration and carefully suspend the cells again.
- Filter the cells through a 70 μm nylon cell strainer to obtain single cell suspension (Figure 4D).
- Seed the cells at 1 x 106 cells in a 35 mm dish after diluting the cells with 2 ml Culture medium and incubate the cells in an incubator at 37 °C for 18 h (Figures 4E and 4F).
- Replace Culture medium in the dish with 2 ml of Maintain Culture medium.
Note: If cells are maintained in the Culture medium for too long, other brain cell types including microglia and astrocyte are likely to grow up. Therefore, Culture medium should be replaced with Maintain culture medium 12-18 h after the cell seeding. - Replace the half of the media in the 35 mm dish with the new 1 ml Maintain culture medium every other day.
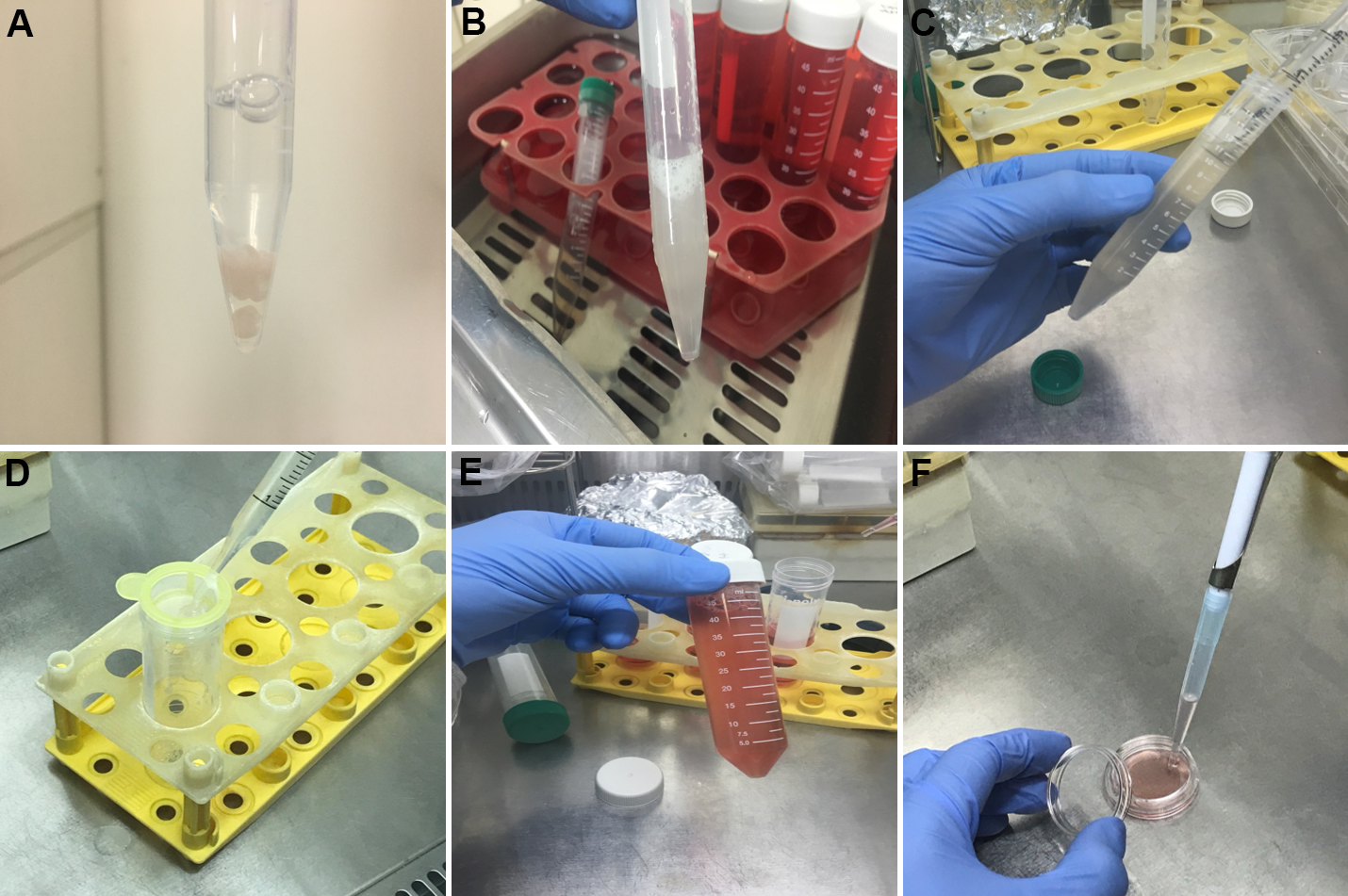
Figure 4. The Process of making single cells of brain tissue and preparing for cell seeding (Steps 10-17) - Grow the cells for at least 7 days after the seeding for experiments (Figure 5).
Note: When you seed the cells, various brain cells (neuron, microglia, astrocytes) are present in the medium. However, after replacement with the Maintain culture medium, only the neuron cells remain specifically because all other types of cells except for the neuron cell are removed by the B-27 constituting the Maintain culture medium.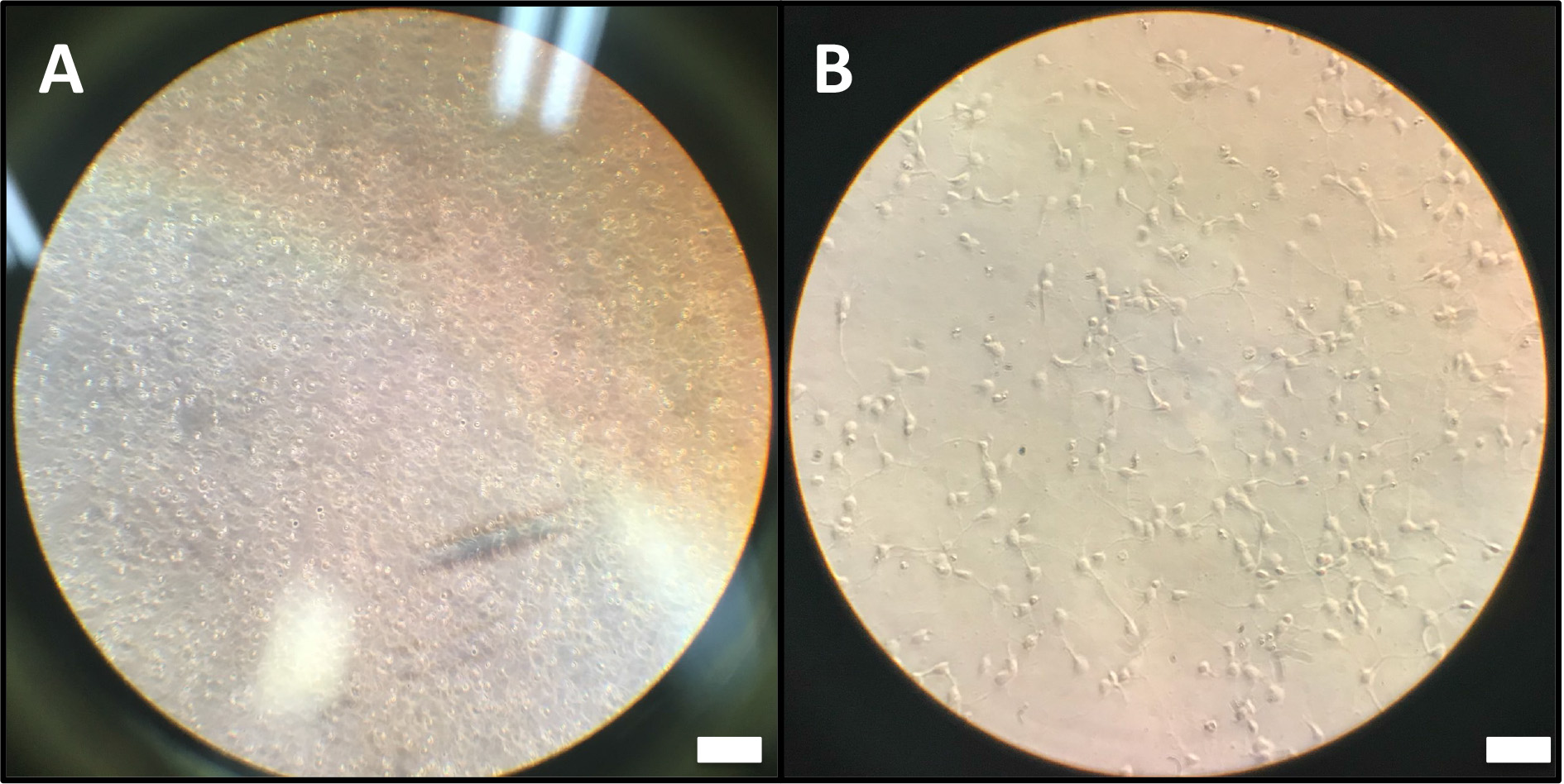
Figure 5. Cell morphology at the 1st day (A) and after 8 days (B). Scale bars, A = 500 μm; B = 200 μm. - Incubate the cells in ROS inducing conditions for about 24 h.
Note: 5 μM of oligomeric Aβ1-42 was added to induce ROS in this protocol, but it may vary depending on your experimental conditions. Aβ1-42 is already known to induce ROS (Andrey et al., 2004, Shelat et al., 2008). - And then treat with 1 μl of CM-H2DCFDA solution (5 mM) or MitoSOXTM Red solution (5 mM) for 20 min.
Note: MitoSOXTM is a specific indicator for mitochondrial superoxide and CM-H2DCFDA is more sensitive to oxidation by H2O2 than superoxide (O2•−). Therefore, even if two chemicals are processed at the same time, they are labeled with different ROS. However, it is difficult to distinguish exactly two types of ROS and analyze fluorescence in the confocal image. We recommend preparing samples separately and conduct each experiment.
Optional: This section provides another option for preparing samples to capture live cell images. If you have cultured cells on a coverslip, carefully put the coverslip into a new 35 mm live cell imaging dish. Put the side of coverslip that cells are attached to the bottom surface. - Replace with the new 2 ml of Maintain culture medium.
Note: Pre-set the condition for taking live cell imaging; 37 °C and 5% CO2 is required because we want to keep the growing conditions for primary neuron cells. - Using confocal microscope, measure the fluorescence mediated by MitoSOXTM (510/580 nm, see Figure 6) or CM-H2DCFDA (495/520 nm, see Figure 7). Since the fluorescence will not last long, it is recommended to measure within 30 min.
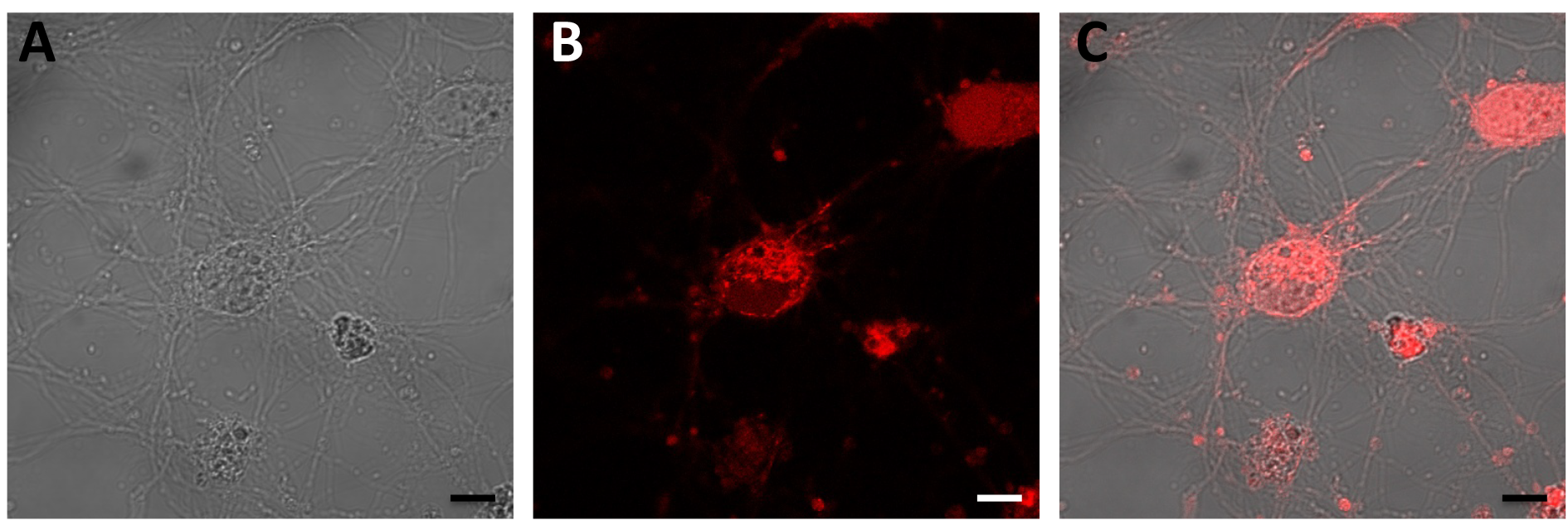
Figure 6. Detection of superoxide in rat primary neuronal cells’ mitochondria with MitoSOXTM Red. Oxidation of MitoSOXTM Red reagent by superoxide produces red fluorescence. A. Bright field rat primary cell images; B. Red fluorescence generated by superoxide; C. Merged image. Scale bars = 50 μm.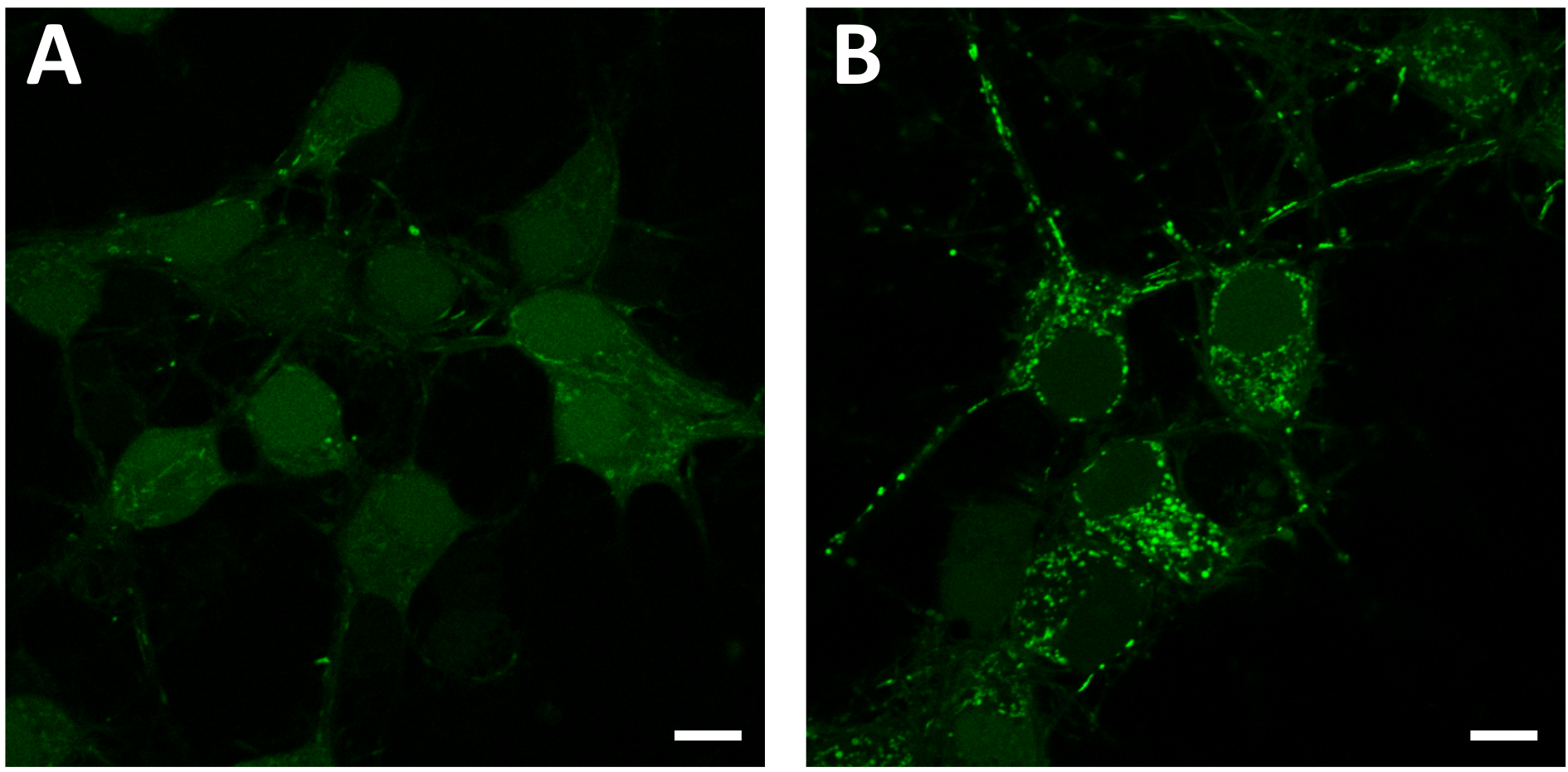
Figure 7. Measurement of ROS in intracellular compartment induced by Aβ in rat primary cells using CM-H2DCFDA. Green fluorescence represents intracellular ROS level of Aβ or vehicle treated sample. A. Vehicle-treated sample; B. 5 μM of Aβ treated sample. Scale bars = 50 μm.
Confocal image measurement setting
You can follow the processes in Figure 8 to measure the level of ROS using confocal microscope:- Run the Zen black edition.
- Click the ‘Smart Setup’ button in ‘Acquisition’ tab to choose the appropriate excitation/emission wavelengths of the dye (MitoSOXTM or CM-H2DCFDA) (Figure 8 A1).
- Set the appropriate excitation/emission wavelengths of the dye and choose the image colors in the ‘Search’ tab (Figure 8C 2).
- Click the ‘Best signal’ button.
- Put the sample on the live cell imaging chamber (Figure 1C).
- Click the ‘Locate’ tab and adjust the focus of confocal microscope (Figure 8B 3).
- After setting the focus of the microscope, click the ‘Set Exposure’ button in ‘Acquisition’ tab.
Note: ‘Set Exposure’ automatically adjusts the detector Gain value (Figure 8A). - Click the ‘Live’ button (Figure 8A).
Note: ‘Live’ performs constant scanning of real-time image. - To get clearer and more accurate images in 'Live' conditions, adjust each value of ‘Gain’, ‘Digital offset’, ‘Digital Gain’, ‘Pinhole’, and laser power (Figure 8D 5). You can also adjust ‘Speed’ and ‘Averaging’ to acquire better images.
Note: In this experiment, ‘Speed’ was set to 7 and ‘Averaging’ number was set to 8. (Figure 9) - Click the ‘Snap’ button to acquire the image (Figure 8A).
Note: All samples should be measured under the same conditions. The measurement method was based on the instructions of the confocal equipment.
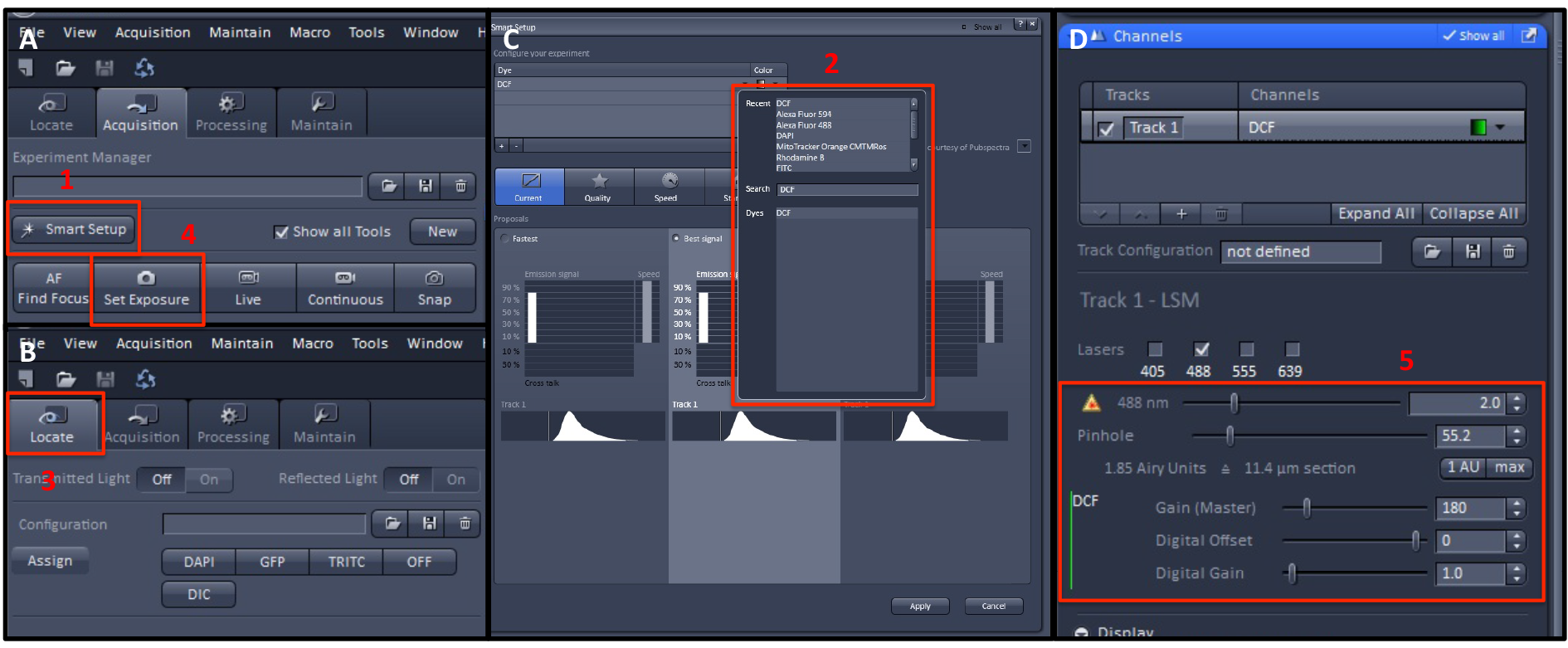
Figure 8. Flow chart of measuring method of ZEN black version of image measurement program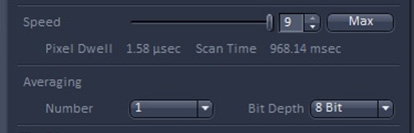
Figure 9. The tab to control 'speed' and 'averaging' in ZEN black version - Run the Zen black edition.
Data analysis
The confocal image that measure the ROS can be used for statistical analysis by quantifying the intensity of fluorescence. This protocol offers two methods.
- Measurement method using basic confocal drive program (see Figure 10).
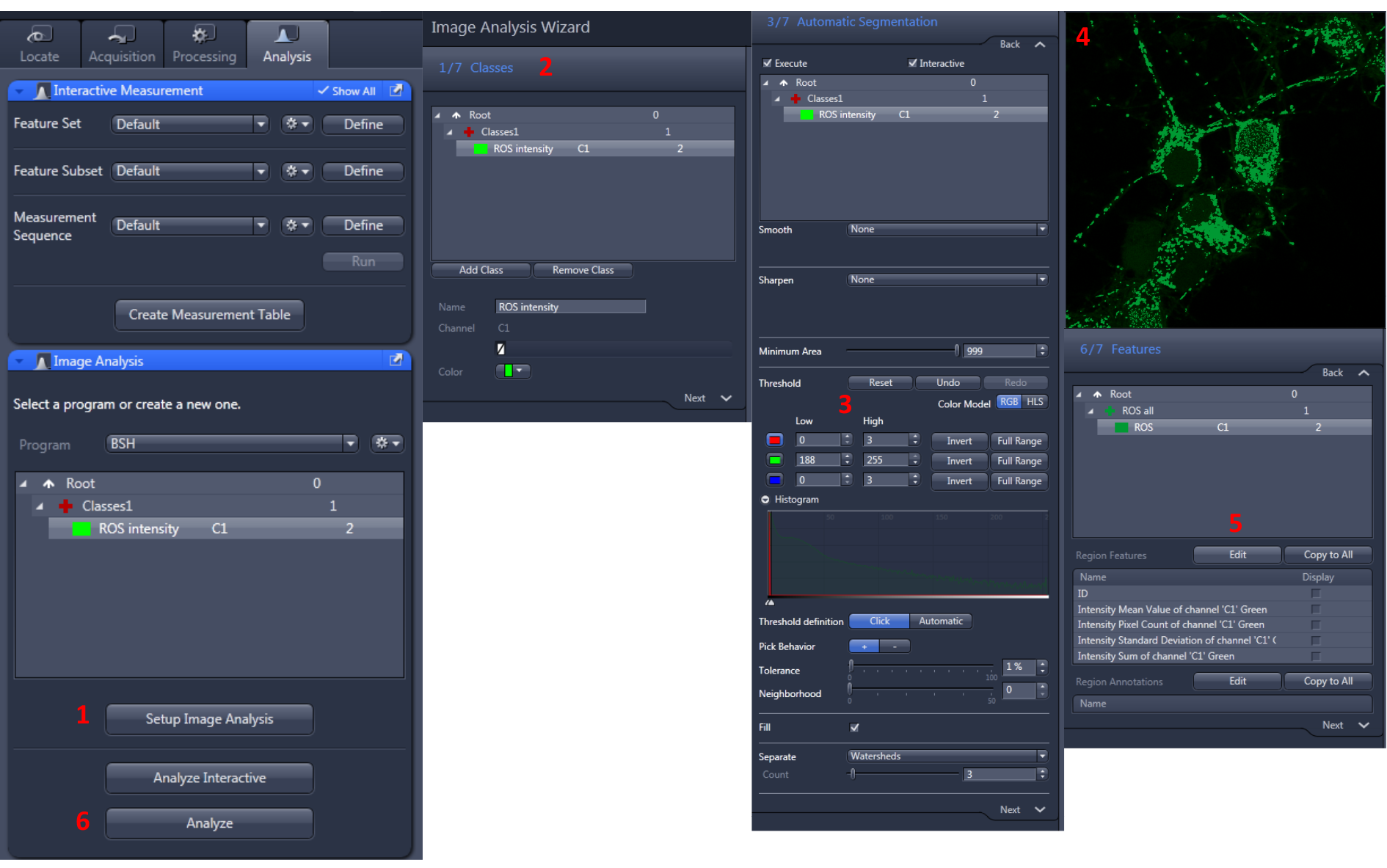
Figure 10. Analysis flowchart of ZEN blue version. Run Zen blue version, an analysis tool provided by Zeiss confocal equipment. Open the image you want to analyze and click the ‘Analysis’ tab. Press the ‘Setup Image Analysis’ button to set the analysis method (see Figure 10-1). Enter the proper analytical condition in order. In particular, set the appropriate threshold value (When you click on the area where CM-H2DCFDA emits fluorescence, the default value is automatically set) in the 3rd step (The analysis setting value must be set up based on the positive control. The settings of all images to be analyzed should be applied equally). Identify the area you want to measure as shown in Figure 10-4. Set the results (Fluorescence mean, Standard deviation, Fluorescence dot number, etc.) you want to obtain and press the ‘Finish’ button. Finally, check the value by pressing the ‘Analysis’ button. - Measurement method using ImageJ (see Figure 11)
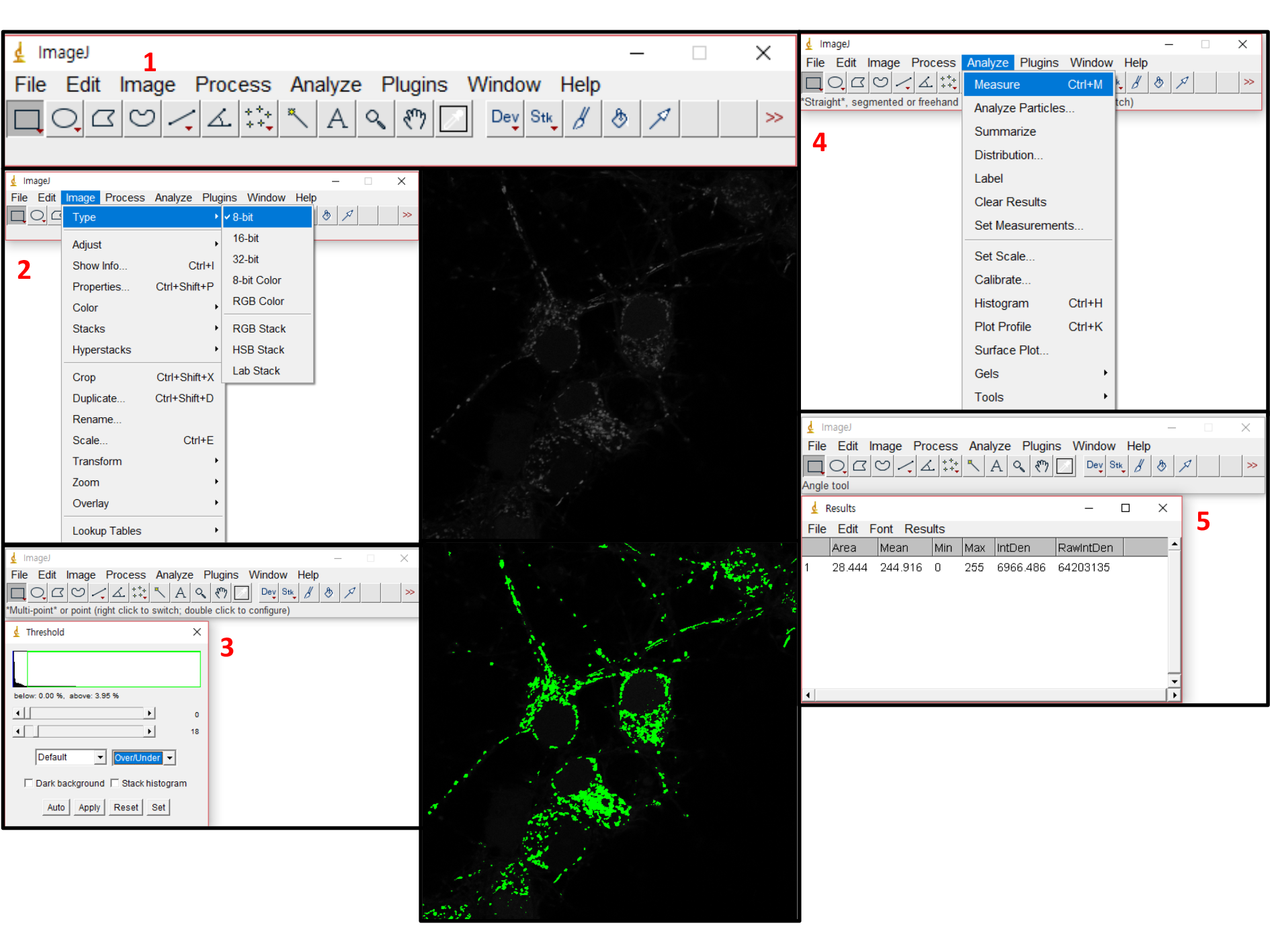
Figure 11. Analysis flowchart of ImageJ. To measure the intensity of fluorescence using ImageJ, several preliminary steps are required. After opening the image you want to analyze, you have to separate the fluorescent area that you want to measure (In flowchart 3, set the range to be measured while adjusting the threshold value. see Figure 11-2, 11-3). Click on the 'Set Measurements' tab as shown in flowchart 4 to set the results you want to obtain. Click the 'Measure' tab and acquire the results.
Notes
- Each experimental group should be treated with MitoSOXTM or CM-H2DCFDA twenty minutes before measuring the fluorescence. You should not treat MitoSOXTM and CM-H2DCFDA in the experimental samples at the same time (Step 19).
- If the cell growing conditions are not maintained, value of ROS measurement will be inaccurate. So, at least 10 minutes of stabilization time should be given before taking confocal images (Step 21).
- When culturing primary neurons, delaying the medium replacement after cell seeding results in a decrease in the percentage of neurons in the cultured cells (Step 16).
Recipes
- CM-H2DCFDA solution (5 mM)
Dissolve 50 μg CM-H2DCFDA (50 μg/1 vial) in 17 μl DMSO - MitoSOXTM Red solution (5 mM)
Dissolve 50 μg MitoSOXTM Red (50 μg/1 vial) in 13 μl DMSO - Poly-D-lysine hydrobromide solution
Poly-D-lysine hydrobromide (5 mg/vial)
50 ml sterile Ultra pure water - Prep medium
PBS 45 ml
5 ml PS - Culture medium
500 ml DMEM
50 ml FBS
5 ml PS - Maintain culture medium
50 ml Neurobasal media
1 ml B-27 supplement
500 μl PS
Acknowledgments
This research was supported by grants (2012R1A5A2A28671860, 2017M3C7A1048268) funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF), the Ministry of Education, Science and Technology, Republic of Korea.
Competing interests
The authors state that they have no conflict of interest to declare.
References
- Andrey, A. Y., Canevari, L. and Duchen, M. R. (2004). β-amyloid peptides induce mitochondrial dysfunction and oxidative stress in astrocytes and death of neurons through activation of NADPH oxidase. J Neurosci 24(2): 565-575.
- Baek, S. H., Park, S. J., Jeong, J. I., Kim, S. H., Han, J., Kyung, J. W., Baik, S. H., Choi, Y., Choi, B. Y., Park, J. S., Bahn, G., Shin, J. H., Jo, D. S., Lee, J. Y., Jang, C. G., Arumugam, T. V., Kim, J., Han, J. W., Koh, J. Y., Cho, D. H. and Jo, D. G. (2017). Inhibition of Drp1 ameliorates synaptic depression, Aβ deposition, and cognitive impairment in an Alzheimer's disease model. J Neurosci 37(20): 5099-5110.
- Batandier, C., Fontaine, E., Keriel, C. and Leverve, X. M. (2002). Determination of mitochondrial reactive oxygen species: methodological aspects. J Cell Mol Med 6(2): 175-187.
- Brieger, K., Schiavone, S., Miller, F. J., Jr. and Krause, K. H. (2012). Reactive oxygen species: from health to disease. Swiss Med Wkly 142: w13659.
- Datta, K., Sinha, S. and Chattopadhyay, P. (2000). Reactive oxygen species in health and disease. Natl Med J India 13(6): 304-310.
- Finkel, T. (2011). Signal transduction by reactive oxygen species. J Cell Biol 194(1): 7-15.
- Forstermann, U. (2008). Oxidative stress in vascular disease: causes, defense mechanisms and potential therapies. Nat Clin Pract Cardiovasc Med 5(6): 338-349.
- Fowler, T. L., Fisher, M. M., Bailey, A. M., Bednarz, B. P. and Kimple, R. J. (2017). Biological characterization of a novel in vitro cell irradiator. PLoS One 12(12): e0189494.
- Forkink, M., Smeitink, J. A., Brock, R., Willems, P. H. and Koopman, W. J. (2010). Detection and manipulation of mitochondrial reactive oxygen species in mammalian cells. Biochim Biophys Acta 1797(6-7): 1034-1044.
- Heneka, M. T., Kummer, M. P. and Latz, E. (2014). Innate immune activation in neurodegenerative disease. Nat Rev Immunol 14(7): 463-477.
- Kirkland, R. A., Saavedra, G. M. and Franklin, J. L. (2007). Rapid activation of antioxidant defenses by nerve growth factor suppresses reactive oxygen species during neuronal apoptosis: evidence for a role in cytochrome c redistribution. J Neurosci 27(42): 11315-11326.
- Khatri, R., Shah, P., Guha, R., Rassool, F. V., Tomkinson, A. E., Brodie, A. and Jaiswal, A. K. (2015). Aromatase inhibitor-mediated downregulation of INrf2 (Keap1) leads to increased Nrf2 and resistance in breast cancer. Mol Cancer Ther 14(7): 1728-1737.
- Liu, Y. H., Weng, Y. P., Lin, H. Y., Tang, S. W., Chen, C. J., Liang, C. J., Ku, C. Y. and Lin, J. Y. (2017). Aqueous extract of Polygonum bistorta modulates proteostasis by ROS-induced ER stress in human hepatoma cells. Sci Rep 7: 41437.
- Nykky, J., Vuento, M. and Gilbert, L. (2014). Role of mitochondria in parvovirus pathology. PLoS One 9(1): e86124.
- Ng, L. F., Gruber, J., Cheah, I. K., Goo, C. K., Cheong, W. F., Shui, G., Sit, K. P., Wenk, M. R. and Halliwell, B. (2014). The mitochondria-targeted antioxidant MitoQ extends lifespan and improves healthspan of a transgenic Caenorhabditis elegans model of Alzheimer disease. Free Radic Biol Med 71: 390-401.
- Polidori, M. C. (2004). Oxidative stress and risk factors for Alzheimer's disease: clues to prevention and therapy. J Alzheimers Dis 6(2): 185-191.
- Ray, P. D., Huang, B. W. and Tsuji, Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 24(5): 981-990.
- Robinson, K. M., Janes, M. S., Pehar, M., Monette, J. S., Ross, M. F., Hagen, T. M., Murphy, M. P. and Beckman, J. S. (2006). Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci U S A 103(41): 15038-15043.
- Singh, D. K., Winocour, P. and Farrington, K. (2011). Oxidative stress in early diabetic nephropathy: fueling the fire. Nat Rev Endocrinol 7(3): 176-184.
- Shelat, P. B., Chalimoniuk, M., Wang, J. H., Strosznajder, J. B., Lee, J. C., Sun, A. Y., Simonyi, A. and Sun, G. Y. (2008). Amyloid beta peptide and NMDA induce ROS from NADPH oxidase and AA release from cytosolic phospholipase A2 in cortical neurons. J Neurochem 106(1): 45-55.
- Trachootham, D., Alexandre, J. and Huang, P. (2009). Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 8(7): 579-591.
- Waris, G. and Ahsan, H. (2006). Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog 5: 14.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Baek, S. H., Cho, Y., Lee, J., Choi, B. Y., Choi, Y., Park, J. S., Kim, H., Sul, J., Kim, E., Park, J. H. and Jo, D. (2018). Intracellular and Mitochondrial Reactive Oxygen Species Measurement in Primary Cultured Neurons. Bio-protocol 8(11): e2871. DOI: 10.21769/BioProtoc.2871.
- Baek, S. H., Park, S. J., Jeong, J. I., Kim, S. H., Han, J., Kyung, J. W., Baik, S. H., Choi, Y., Choi, B. Y., Park, J. S., Bahn, G., Shin, J. H., Jo, D. S., Lee, J. Y., Jang, C. G., Arumugam, T. V., Kim, J., Han, J. W., Koh, J. Y., Cho, D. H. and Jo, D. G. (2017). Inhibition of Drp1 ameliorates synaptic depression, Aβ deposition, and cognitive impairment in an Alzheimer's disease model. J Neurosci 37(20): 5099-5110.
Category
Neuroscience > Cellular mechanisms > Intracellular signalling
Neuroscience > Nervous system disorders > Cellular mechanisms
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link