- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ectopic Gene Expression in Macrophages Using in vitro Transcribed mRNA
Published: Vol 8, Iss 10, May 20, 2018 DOI: 10.21769/BioProtoc.2857 Views: 8512
Reviewed by: Alka MehraGal HaimovichRan Chen

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Vitro Analysis of Stalled Ribosomes using Puromycin Incorporation
MaKenzie R. Scarpitti and Michael G. Kearse
Aug 20, 2023 3373 Views

Immunoprecipitation of Reporter Nascent Chains from Active Ribosomes to Study Translation Efficiency
Roberta Cacioppo and Catherine Lindon
Sep 20, 2023 2148 Views

Metabolic RNA Labeling and Translating Ribosome Affinity Purification for Measurement of Nascent RNA Translation
Hirotatsu Imai and Akio Yamashita
Oct 20, 2024 2544 Views
Abstract
Macrophages are immune cells that contribute to host defense through various mechanisms including phagocytosis and antigen presentation. Their antimicrobial capacity is subverted by clinically important intracellular pathogens such as Mycobacterium tuberculosis. The study of host-pathogen interactions using these cells is therefore of considerable interest. Such studies often seek to express tagged proteins to characterize their activities, localizations, and protein-protein interactions. Here, we describe a robust method for transient protein expression in macrophages using mRNA lipoplex transfections.
Keywords: mRNA transfectionBackground
Typical methods to achieve protein expression, including transfecting DNA using cationic polymers, nucleofection, and viral transduction (Zhang et al., 2009), are particularly difficult in macrophages, as these cells have a potent immune response to various danger signals such as cytosolic DNA. Therefore, conventional methods for exogenous gene delivery result in poor transfection efficiency and cell death. We reasoned that transfecting mRNA, instead of DNA, would be a better alternative to achieve protein expression in macrophages, as suggested by recent reports (Van De Parre et al., 2004; McLenachan et al., 2013). We were able to achieve high transfection efficiencies without loss of macrophage viability (Koster et al., 2017). Our method does not require expensive equipment and can be adapted for expressing exogenous and endogenous proteins.
Materials and Reagents
- μ-Plate 96 Well (ibidi, catalog number: 89626 )
- Sterile RNase-free microfuge tubes (Thermo Fischer Scientific, catalog number: AM12400 )
- DNase and RNase free sterile pipet tips (e.g., Sorenson Bioscience, catalog number: 10350 )
- Gloves (e.g., powder free nitrile gloves, MICROFLEX, catalog number: XC-310 )
- Macrophages
Macrophage cell types that can be used are cell lines such as RAW 264.7 (ATCC, catalog number: TIB-71 ) and primary macrophages such as C57BL/6 bone marrow-derived macrophages (BMDMs). - Cell culture medium
RAW 294.7 cell lines were maintained in Dulbecco Modified Eagle Medium (DMEM, Thermo Fisher Scientific, GibcoTM, catalog number: 11965 ) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Thermo Fisher Scientific, GibcoTM, catalog number: 10082147 ). - CleanCapTM EGFP mRNA (TriLink BioTechnologies, catalog number: L-7601 )
- TransIT®-mRNA Transfection Kit (Mirus Bio, catalog number: MIR 2250 )
- OptiMEM medium (Thermo Fisher Scientific, GibcoTM, catalog number: 31985062 )
- RNaseZapTM (Sigma-Aldrich, catalog number: R2020 )
- mMESSAGE mMACHINE T7 Ultra Kit (Thermo Fisher Scientific, AmbionTM, catalog number: AM1345 )
- MEGAClearTM Transcription Clean-Up Kit (Thermo Fisher Scientific, AmbionTM, catalog number: AM1908 )
- Molecular biology grade water (Corning, catalog number: 46-000-CI )
- Fetal bovine serum (Thermo Fisher Scientific, GibcoTM, catalog number: 16140071 )
- Penicillin and streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- BglII restriction endonuclease (New England Biolabs, catalog number: R0144S )
- NEBuffer 3.1 (New England Biolabs, catalog number: B7203S )
- Antarctic Phosphatase Reaction Buffer (New England Biolabs, catalog number: B0289S )
- Antarctic Phosphatase (New England Biolabs, catalog number: M0289S )
- QIAquick PCR Purification Kit (QIAGEN, catalog number: 28106 )
- GatewayTM pcDNATM-DEST47 vector (Thermo Fischer Scientific, catalog number: 12281010 )
- Agarose (LE Agarose, GeneMate, catalog number: E-3120-500 )
- 10x MOPS buffer (Fischer Scientific, catalog number: BP2900500 )
- Ethidium bromide (Sigma-Aldrich, catalog number: E1510 )
- RNA Millennium Markers (Thermo Fisher Scientific, AmbionTM, catalog number: AM7150 )
- RNA gel loading dye (Thermo Fischer Scientific, catalog number: R0641 )
- Formaldehyde solution (37%, Sigma-Aldrich, catalog number: 252549 )
- Plasmid linearization reaction mix (see Recipe 1)
- EGFP mRNA transfection reaction mix (see Recipe 3)
- Poly(A) tailing reaction mix (see Recipe 4)
- RNA denaturing gel electrophoresis (see Recipe 5)
- mRNA in vitro transcription reaction mix (see Recipe 6)
- DMEM complete medium (see Recipe 7)
Equipment
- P20 Pipetman (Gilson, catalog number: F123600 )
- P200 Pipetman (Gilson, catalog number: F123601 )
- P1000 Pipetman (Gilson, catalog number: F123602 )
- Multi-channel pipette (Eppendorf, catalog number: 3125000044 )
- Tissue culture CO2 incubator
- Tissue culture hood
- Water bath
- -20 °C freezer
- Epifluorescence microscope (e.g., Nikon, model: Eclipse Ti-E , equipped with 60x; Plan-Apochromat, NA 1.4 oil immersion objective, Ti Z drive, high-resolution monochrome charge-coupled device (CCD) digital camera; Photometric Cool SNAP HQ2 and appropriate filter sets for DAPI, FITC and TexasRed channel)
- Eppendorf Thermomixer® C (Eppendorf, model: ThermoMixer® C , catalog number: 5382000023)
- NanoDrop Spectrophotometer
- Agarose gel electrophoresis system
Software
- Nikon Imaging Software-Elements Advanced Research (NIS-Elements) version 4.40
Procedure
- In vitro mRNA transcription
Note: There are multiple methods to obtain mRNA for the gene of interest. One can use PCR amplified DNA or linearized plasmid DNA that contains an RNA polymerase binding site (McLenachan et al., 2013). Alternatively, mRNA can be purchased commercially. Here, we describe in vitro mRNA transcription using the plasmid, pcDNA-DEST47-CpsA-GFP, which has a T7 promoter and polyadenylation sites (Koster et al., 2017).
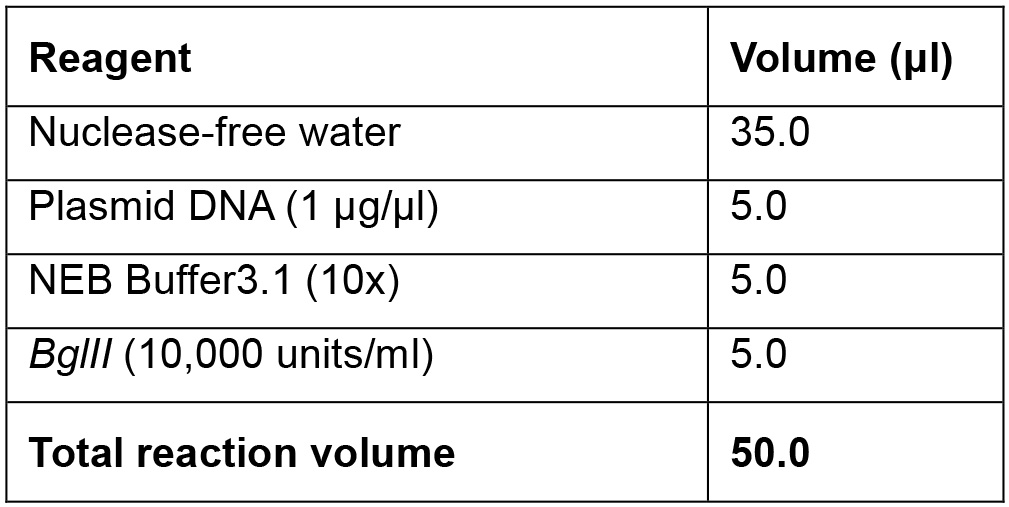
Linearization of plasmid DNA- Linearize the plasmid DNA using the restriction endonuclease BglII in a total reaction volume of 50 μl, at 37 °C for 2 h (see Recipe 1 below).
Notes:- One may choose from a variety of restriction sites present in the plasmid for linearization. For example, XmaI can also be used instead of BglII in this case. Plasmid linearization should preferably be done using a restriction enzyme that results in blunt ends or 5’ overhangs downstream of the gene of interest. Temperature and duration of incubation will depend upon the restriction enzyme used.
- Start with enough amount of plasmid DNA to account for sample loss. For example, 3 μg of plasmid yielded ~2.5 μg of final purified linear product.
- Heat inactivation of the restriction enzyme must be done after completion of plasmid digestion. Temperature/time will depend upon enzyme used. In the case where it is not possible, such as in case of BglII, proceed to the next step immediately.
- One may choose from a variety of restriction sites present in the plasmid for linearization. For example, XmaI can also be used instead of BglII in this case. Plasmid linearization should preferably be done using a restriction enzyme that results in blunt ends or 5’ overhangs downstream of the gene of interest. Temperature and duration of incubation will depend upon the restriction enzyme used.
- To the above solution, add Antarctic Phosphatase reaction buffer and Antarctic Phosphatase enzyme (1 unit per μg plasmid) to make a final volume of 60 μl (see Recipe 2 below).
Note: This step is important to dephosphorylate the 5’ and 3’ phosphorylated ends of the linearized plasmid and prevent re-ligation. - Incubate the above reaction mixture for 1 h at 37 °C.
- Heat-inactivate the Antarctic Phosphatase at 80 °C for 2 min.
- Purify the linearized plasmid DNA from the above reaction mixture using the PCR Purification kit (QIAquick PCR Purification Kit, QIAGEN) and quantify the product by taking absorbance measurement at 260 nm.
- Confirm plasmid linearization by comparison with an uncut plasmid using 1% agarose gel electrophoresis (Figure 1).
Note: It is important to confirm that the plasmid has been linearized as the presence of uncut plasmid DNA will significantly decrease the yield of desired mRNA product. If there is uncut plasmid contamination, the linearized plasmid band can be purified from the agarose gel using the Qiaquick Gel Extraction Kit (QIAGEN). Alternatively, one can re-optimize conditions for Step A1.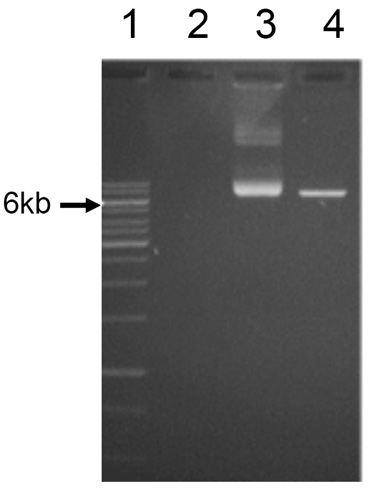
Figure 1. Agarose gel with 1 kb DNA ladder (#1), uncut and linear plasmid (#3, 4)
Notes:- Thaw the plasmid DNA and mRNA transcription kit reagents according to the manufacturer’s protocol. Vortex and spin down the vials to ensure homogenous solutions.
- The kit used here describes reaction steps for synthesizing mRNA in the range of 300 bases to 5 kb. However, this protocol has been optimized for ~8 kb product.
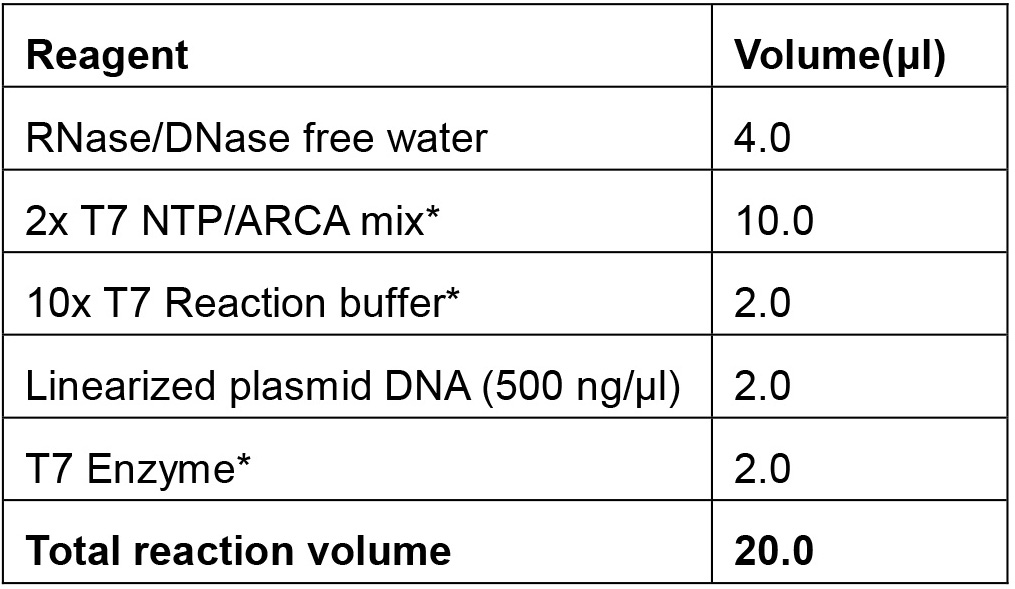
- Assemble a 20 μl transcription reaction (see Recipe 3 below) by diluting linear plasmid, 2x NTP/ARCA buffer, 10x reaction buffer, and T7 enzyme in nuclease-free water. The reaction mix should be made at room temperature.
Note: The kit adds a novel cap analog called anti-reverse cap analog (ARCA) which ensures correct mRNA capping orientation. - Mix the contents well by gently pipetting up and down.
- Incubate the reaction mixture at 37 °C for 2 h.
- Add 1 μl of Turbo DNase to the above reaction mixture and incubate for 15 min at 37 °C.
Note: DNase addition is important to eliminate DNA contamination from the end product.
- To the above 20 μl transcription reaction, add 5x E-PAP buffer, MnCl2 solution, ATP solution and nuclease-free water to make a final volume of 96 μl (see Recipe 4 below).
- Take out a 2.5 μl aliquot of this reaction mixture and store it for later use in Step A18.
- Add 4 μl of E-PAP enzyme and mix gently by pipetting up and down.
- Incubate the reaction mixture at 37 °C for 45 min.
- Purify the mRNA product using the MEGAClear Kit according to the manufacturers’ instructions.
- Quantify mRNA by taking absorbance at 260 nm.
Notes:- Aliquot the transcribed mRNA in separate RNase/DNase free tubes. They can be stored at -20 °C for several months or -80 °C for years.
- Using the above method 60 µg mRNA can be obtained from 1 µg linear plasmid.
- Aliquot the transcribed mRNA in separate RNase/DNase free tubes. They can be stored at -20 °C for several months or -80 °C for years.
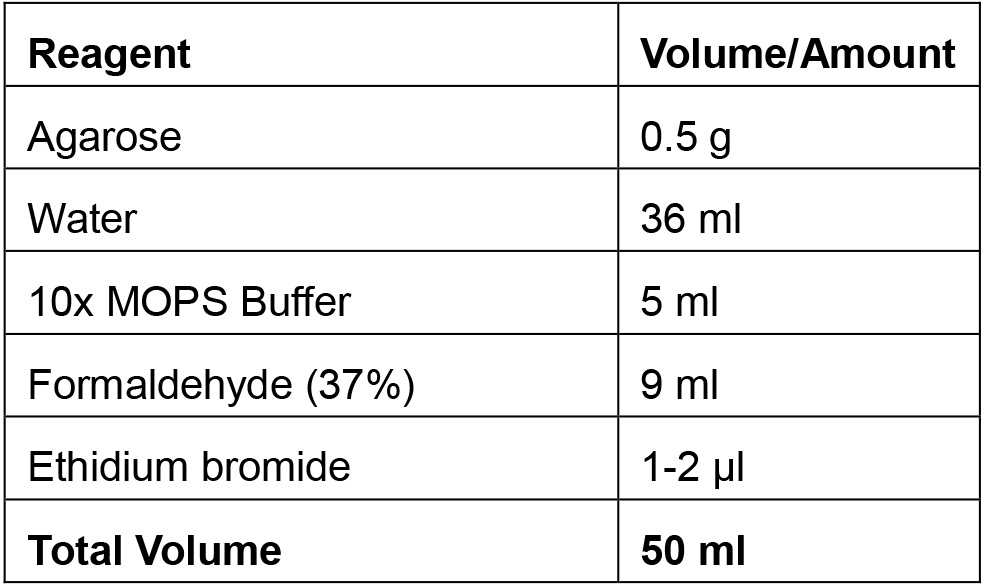
- RNA denaturing gel electrophoresis should be performed to assess the quality of the transcribed mRNA. To prepare 50 ml of 1% agarose gel, add 0.5 g agarose in 36 ml double-distilled water, and boil. Add 5 ml 10x MOPS buffer and 9 ml formaldehyde (37%). Add ethidium bromide at a final concentration of 0.5 μg/ml (see Recipe 5 below).
Notes:- Formaldehyde should be handled inside a chemical fume hood. Do not boil agarose solution with formaldehyde.
- Ethidium bromide is a known mutagen and therefore careful handling is important. One can also use safer alternatives such as SYBR Safe dye (Invitrogen).
- Formaldehyde should be handled inside a chemical fume hood. Do not boil agarose solution with formaldehyde.
- Dilute the RNA marker and the RNA samples from Steps A12 and A16 in 2x RNA loading dye and boil at 75 °C for 10 min.
- Load the RNA samples and run the gel in 1x MOPS buffer at ~5 volts/cm. Visualize RNA bands under UV illumination (Figure 2).
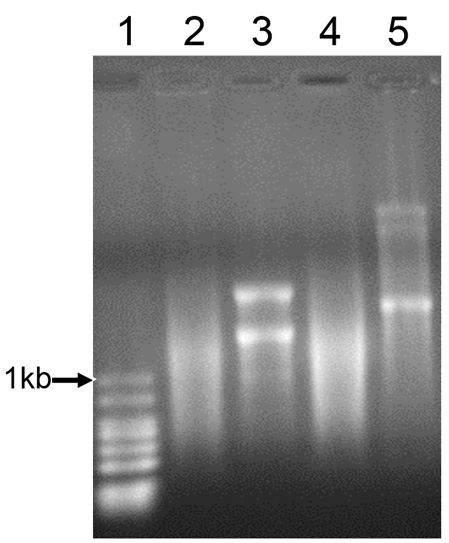
Figure 2. Agarose gel with RNA ladder (#1), CAT-GFP mRNA before (#2) and after poly(A) tailing (#3) and CpsA-GFP mRNA before (#4) and after poly(A) tailing (#5) - Transfect RAW 264.7 macrophages with the in vitro transcribed mRNA as described in Procedure B.
- Samples can be analyzed 24-72 h later. We performed phagocytosis assays 24 h after transfection (Koster et al., 2017).
Notes:- It is advised to setup transfection with EGFP mRNA in parallel as a positive control for transfection efficiency. Boost and Transfection Reagents alone, without mRNA, is not a useful negative experimental control. Uncomplexed reagents may result in cell toxicity. An appropriate experimental control will depend upon the specific study. We used the chloramphenicol resistance gene (CAT) in the pcDNATM-DEST47 vector to make CAT-GFP mRNA as a control (Figure 2).
- This method results in transient protein expression in macrophages. The duration of protein expression will depend upon the turnover of the specific protein analyzed. The user may re-transfect the macrophages, typically after 48 h, if longer experimental durations are desired.
- It is advised to setup transfection with EGFP mRNA in parallel as a positive control for transfection efficiency. Boost and Transfection Reagents alone, without mRNA, is not a useful negative experimental control. Uncomplexed reagents may result in cell toxicity. An appropriate experimental control will depend upon the specific study. We used the chloramphenicol resistance gene (CAT) in the pcDNATM-DEST47 vector to make CAT-GFP mRNA as a control (Figure 2).
- Linearize the plasmid DNA using the restriction endonuclease BglII in a total reaction volume of 50 μl, at 37 °C for 2 h (see Recipe 1 below).
- Standardization of macrophage transfections using EGFP mRNA
- Seed RAW 264.7 macrophages in a 96-well black plate (µ-Plate 96 well, ibidi, GmbH) at a density of 15,000 cells/well. Incubate the cells in a humidified incubator at 37 °C with 5% CO2 atmosphere.
Note: The protocol can be adapted to other well formats as well. Macrophage seeding density will depend on the replication rate of the cells as well as the duration of the experiment. - The next day, transfect macrophages with EGFP mRNA using the Mirus TransIT mRNA transfection kit. Start by putting the contents of the mRNA transfection kit (Boost Reagent and Transfection Reagent) and OptiMEM at room temperature.
Note: Do not place Boost Reagent or Transfection Reagent on ice or in the water bath. - Thaw EGFP mRNA on ice.
Notes:- To preserve mRNA integrity, always thaw mRNA on ice and avoid repeated freeze-thaws. It is advisable to make aliquots of the mRNA solution.
- One must perform RNA work with extreme caution. Clean the workplace and spray RNaseZap to prevent RNase contamination. Always wear gloves and change them regularly. Always use nuclease-free tips and tubes for RNA work.
- To preserve mRNA integrity, always thaw mRNA on ice and avoid repeated freeze-thaws. It is advisable to make aliquots of the mRNA solution.
- Before transfecting the macrophages, remove the culture medium and replace with 150 μl of fresh complete medium. Return the cells to the incubator.
- Prepare 50 μl per well of the mRNA transfection mix (see Recipe 6 below) by diluting 2 μl EGFP mRNA (100 μg/ml stock solution) in 40 μl OptiMEM. Mix well by gently pipetting up and down.
Note: The volumes described are for the transfection of 200 ng EGFP mRNA. It is advised to use a range of mRNA amounts such as 100, 200 and 500 ng for standardization according to the experimental cell types and conditions used. - Add Boost Reagent to the above solution at 2 μl per μg mRNA. Mix well by gently pipetting up and down.
- Add Transfection Reagent to the above solution at 2 μl per μg mRNA. Mix well by gently pipetting up and down.
Note: Boost and transfection reagents can be diluted in OptiMEM before starting the protocol. However, they must be diluted freshly and should not be stored for later use. A ratio of 2 μl Boost/Transfection reagent per μg mRNA was used in this protocol, however, one may modify this ratio depending on the cell type and tissue culture plate format. - Incubate the transfection mix at room temperature for 3-5 min.
Note: This is a critical step. Incubating longer than 5 min may decrease transfection efficiency. - Drop 50 μl per well of the mRNA transfection solution into the macrophage culture. Rock the plate back and forth, side to side very gently to ensure proper mixing.
- Incubate transfected macrophages in the tissue culture incubator overnight.
- The next day, observe the GFP transfected cells using a fluorescent microscope, using filter sets for excitation at 488 nm and emission at 530 nm (FITC channel).
Note: Cell viability can be assessed using standard methods such as staining with Alamar blue or trypan blue dyes (Thermo Fischer Scientific). For the protocol described here, cell viability declined when 500 ng mRNA was used for transfection. In addition, lab specific assays of immune activation can be performed.
- Seed RAW 264.7 macrophages in a 96-well black plate (µ-Plate 96 well, ibidi, GmbH) at a density of 15,000 cells/well. Incubate the cells in a humidified incubator at 37 °C with 5% CO2 atmosphere.
Data analysis
To quantify the EGFP signal in the fluorescent cells (Step B11, Figure 3), a basic workflow is described below using NIS-Elements version 4.40.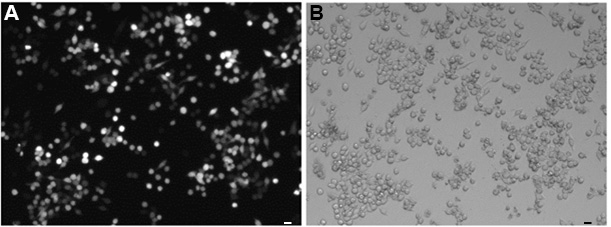
Figure 3. Expression of EGFP in RAW264.7 macrophages. Cells were transfected with 200 ng of EGFP mRNA. A. The EGFP fluorescent cells were visualized using a fluorescent microscope. B. A bright-field image of the cells. Scale bars = 20 μm.
- Open the image of the EGFP positive cells in the software and click on the Region of Interest (ROI) icon on the right side of the image border. Choose an option to draw an ROI around the cells in the image. As examples, I have selected 3 ROIs having different GFP intensities (Figure 4).
Note: Most image analysis software have similar options and one can use those for image analysis as well. For example, ImageJ has ‘rectangular’, ‘oval’ and ‘polygonal’ selection options.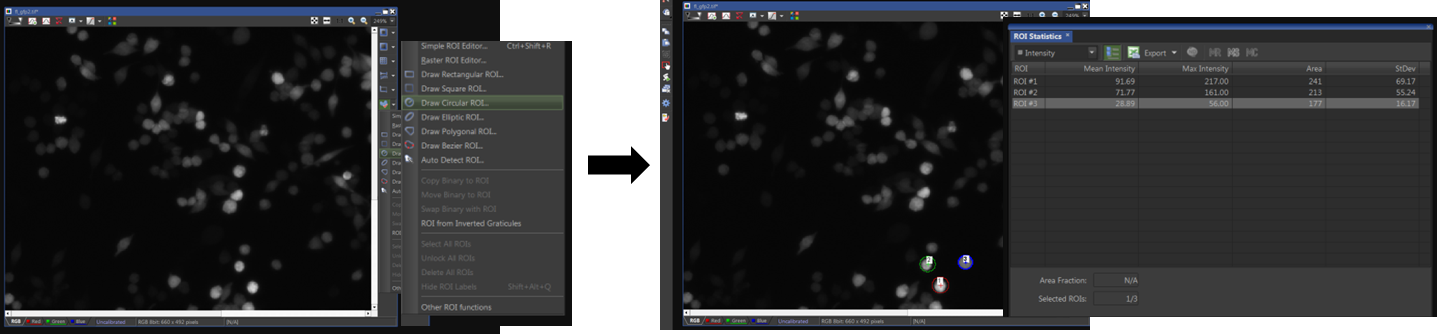
Figure 4. Selection of ROIs for EGFP intensity analysis. An ROI was drawn around individual cells for GFP intensity analysis. - Right-click on the NIS-Elements desktop and select ‘Analysis Controls’. Select the option ‘ROI Statistics’.
- The software will compute the mean fluorescence intensity (MFI) for each selected ROI (Figure 4). Select the option ‘Export’ in the ROI statistics window to export your data out in an Excel sheet.
- Apply the same method to obtain MFI of cells in the untransfected sample. Calculate the average of this set of values. This will estimate the autofluorescence.
- Calculate transfection efficiency using the formula:
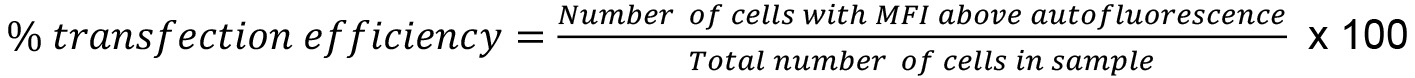
Note: The transfection efficiency achieved using describe protocol was 60-70%.
Recipes
- Plasmid linearization reaction mix
- Antarctic Phosphatase reaction mix

- mRNA in vitro transcription reaction mix
*provided in the mMESSAGE mMACHINE T7 Ultra Kit - Poly(A) tailing reaction mix

*provided in the mMESSAGE mMACHINE T7 Ultra Kit - RNA denaturing gel electrophoresis
- EGFP mRNA transfection reaction mix
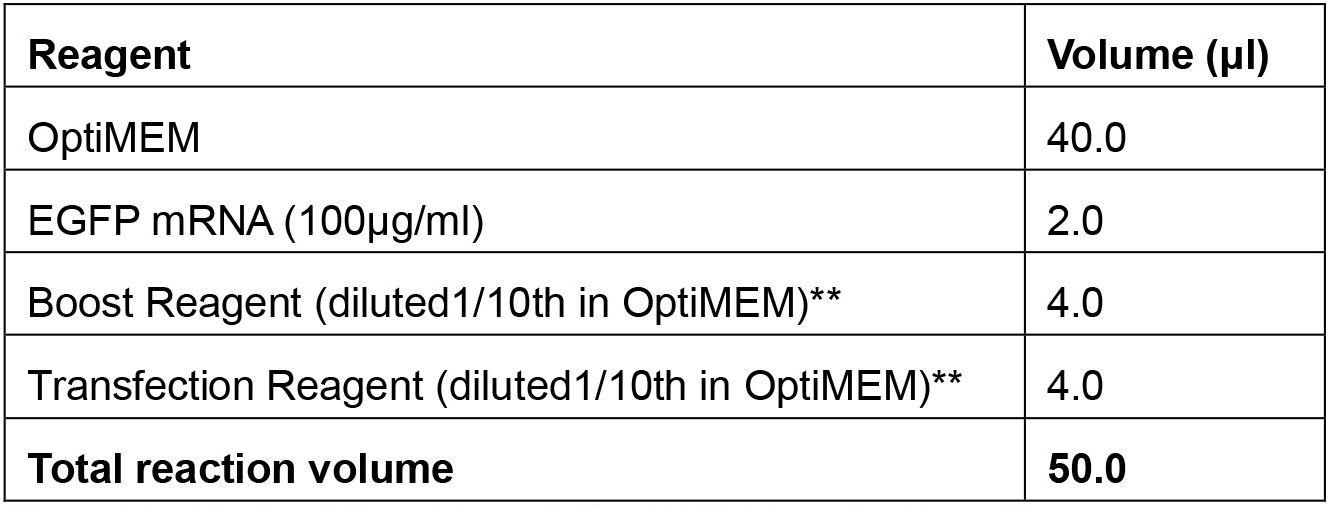
**provided in the Mirus TransIT®-mRNA Transfection Kit. - DMEM complete medium

The medium should be sterile for use in tissue culture. One may maintain cells in complete medium with penicillin and streptomycin (Gibco).
Acknowledgments
This work was funded by the NIH/NIAID (R01 AI087682, 1R01 AI130454, and R21 AI128427), Potts Memorial Foundation, and Washington University in St Louis School of Medicine. The authors have no conflicts of interest. This protocol was used for studies reported in Koster et al. (2017).
References
- Koster, S., Upadhyay, S., Chandra, P., Papavinasasundaram, K., Yang, G., Hassan, A., Grigsby, S. J., Mittal, E., Park, H. S., Jones, V., Hsu, F. F., Jackson, M., Sassetti, C. M. and Philips, J. A. (2017). Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proc Natl Acad Sci U S A 114(41): E8711-E8720.
- McLenachan, S., Zhang, D., Palomo, A. B., Edel, M. J. and Chen, F. K. (2013). mRNA transfection of mouse and human neural stem cell cultures. PLoS One 8(12): e83596.
- Van De Parre, T. J. L., Martinet, W., Schrijvers, D. M., Herman, A. G. and De Meyer, G. R. Y. (2004). mRNA but not plasmid DNA is efficiently transfected in murine J774A.1 macrophages. Biochem Biophys Res Commun 327: 356-360.
- Zhang, X., Edwards, J. and Mosser, D. (2009). The expression of exogenous genes in macrophages: obstacles and opportunities. In: Reiner, N. (Ed.) Macrophages and Dendritic Cells. Methods in Molecular Biology (Methods and Protocols). Vol 531. Humana Press.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chandra, P. and Philips, J. A. (2018). Ectopic Gene Expression in Macrophages Using in vitro Transcribed mRNA. Bio-protocol 8(10): e2857. DOI: 10.21769/BioProtoc.2857.
Category
Molecular Biology > RNA > mRNA translation
Molecular Biology > RNA > Transfection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










