- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mammalian Cell-derived Vesicles for the Isolation of Organelle Specific Transmembrane Proteins to Conduct Single Molecule Studies
Published: Vol 8, Iss 9, May 5, 2018 DOI: 10.21769/BioProtoc.2825 Views: 7497
Reviewed by: Rahul SrinivasanTalita Diniz Melo HanchukAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Refined Protocol for Identifying Citrulline-specific Monoclonal Antibodies from Single Human B Cells from Rheumatoid Arthritis Patient Material
Khaled Amara [...] Caroline Grönwall
Sep 5, 2019 6991 Views

Isolation and Characterization of Exosomes from Mouse Feces
Chunhua Yang [...] Didier Merlin
Apr 20, 2020 7636 Views
Abstract
Cell-derived vesicles facilitate the isolation of transmembrane proteins in their physiological membrane maintaining their structural and functional integrity. These vesicles can be generated from different cellular organelles producing, housing, or transporting the proteins. Combined with single-molecule imaging, isolated organelle specific vesicles can be employed to study the trafficking and assembly of the embedded proteins. Here we present a method for organelle specific single molecule imaging via isolation of ER and plasma membrane vesicles from HEK293T cells by employing OptiPrep gradients and nitrogen cavitation. The isolation was validated through Western blotting, and the isolated vesicles were used to perform single molecule studies of oligomeric receptor assembly.
Keywords: Single moleculeBackground
A large number of transmembrane proteins are formed through the assembly of multiple subunits leading to complicated oligomeric structures that can often exist in multiple stoichiometries. Understanding how changes in assembly alter trafficking and localization within different organelles is essential to determining a protein’s physiological role and the connection to diseases associated with maturation and transport. Single molecule approaches can provide a better understanding of the assembly of oligomeric proteins by directly measuring their stoichiometry (Ulbrich and Isacoff, 2007; Richards et al., 2012). This approach avoids ensemble averaging which provides the average state of all the stoichiometries (Walter and Bustamante, 2014). Single molecule studies have recently been employed to understand the structural and functional properties of macromolecules including conformational dynamics (Tan et al., 2014), ion channel gating (Wang et al., 2016), ligand-receptor interaction (Moonschi et al., 2015), and stoichiometric assembly (Ulbrich and Isacoff, 2007; Moonschi et al., 2015). To conduct single molecule studies, it is imperative to isolate single receptors in a supporting bilayer. Separation from the cellular environment is often necessary because the endogenous concentration of membrane proteins in mammalian cells is typically much higher than conditions compatible with single molecule studies (Richards et al., 2012). Additionally, live cell single molecule studies suffer from a number of disadvantages including high levels of auto-fluorescence, limited fluorophore brightness and the mobility of membrane proteins on the cell surface (Andersson et al., 1998; Lippincott-Schwartz et al., 1999). Isolation strategies using artificial bilayers such as liposomes are also problematic as they include an intermediate step where the protein is stabilized into a detergent solution which poses a threat to the structural integrity of the transmembrane protein. Membrane-derived vesicles enable the protein to remain embedded in its physiological membrane maintaining its structural and functional integrity. These nanoscale vesicles have been employed to study stoichiometric assembly of multimeric proteins, to probe ligand receptor interactions, and to understand the effect of ligands on the assembly of nicotinic receptors (Fox et al., 2015; Moonschi et al., 2015; Fox-Loe et al., 2017).
Transmembrane proteins are synthesized and assembled in the endoplasmic reticulum and then trafficked to the plasma membrane. Oligomeric proteins with multiple non-identical subunits can often be assembled with different subunit stoichiometries potentially leading to different trafficking and functional properties (Grady et al., 2010). For instance, α4β2 nicotinic receptors are pentameric receptor with two possible stoichiometric assemblies: (α4)2(β2)3 and (α4)3(β2)2. It has been hypothesized that cellular machinery preferentially traffics the high sensitivity isoform (α4)2(β2)3 over the other isoform from the ER to the plasma membrane. It has also been hypothesized that nicotine alters the assembly of this receptor from the low sensitivity to high sensitivity isoform in the ER leading to higher levels of the preferentially trafficked assembly in both the ER and plasma membrane (Lester et al., 2009; Henderson and Lester, 2015). Single molecule studies of proteins from specific organelles enable the correlation of changes in structural assembly of the nicotinic receptors to changes in trafficking and assembly.
Here we present a method which can isolate single transmembrane proteins into the ER and plasma membrane derived vesicles using nitrogen cavitation and an OptiPrep gradient. The ER and plasma membrane derived vesicles exhibit different densities because they contain different phospholipids and associated proteins. ER originated vesicles are much denser than those obtained from the plasma membrane. The OptiPrep gradient was selected because of its superior ability to maintain isosmotic pressure independent of the density of the gradient used to isolate cellular organelles and subcellular vesicles (Graham et al., 1994). We applied this method to study ligand induced changes in the assembly of nicotinic receptors in the ER as well as the plasma membrane and validated our method with Western blotting and single molecule step-wise photobleaching. We believe this method can be applied for virtually any type of transmembrane proteins to conduct single molecule studies and to understand organelle-specific structural and functional properties.
Materials and Reagents
- Cell culture flasks (Greiner Bio One International, catalog number: 658175 )
- 50 ml centrifuge tubes (VWR, catalog number: 89039-658 )
- Ultra-Clear ultracentrifuge tubes (Beckman Coulter, catalog number: 344061 )
- Four 5-ml Serological pipettes (VWR, catalog number: 89130-896 )
- One 9-inch-flint-glass Pasteur pipette (VWR, catalog number: 14672-380 )
- 15 ml centrifuge tubes (VWR, catalog number: 89039-666 )
- 1.5-ml tubes
- Wine cork
- Razor blades (VWR, catalog number: 55411-050 )
- Two ice buckets (VWR, catalog number: 10146-202 )
- HEK293T cells (Sigma-Aldrich, catalog number: 85120602-1VL )
- Deionized water
- PBS buffer (VWR, catalog number: 97062-948 )
- Versene solution (Thermo Fisher Scientific, GibcoTM, catalog number: 15040066 )
- Nitrogen gas
- OptiPrep (60% solution, Sigma-Aldrich, catalog number: D1556-250ML )
- Anti-calnexin (Abcam, catalog number: ab92573 )
- Anti-sodium potassium ATPase (Abcam, catalog number: ab76020 )
- Mouse anti-Rabbit secondary antibody (Santa Cruz Biotechnology, catalog number: sc-2357 )
- ClarityTM Western ECL Substrate (Bio-Rad Laboratories, catalog number: 1705060 )
- Tris-HCl (Sigma-Aldrich, Roche Diagnostics, catalog number: 10812846001 )
- Sodium chloride (NaCl) (Fisher Scientific , catalog number: BP358-1 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Fisher Scientific, catalog number: BP214-500 )
- Calcium chloride (CaCl2) (Fisher Scientific, catalog number: C614-500 )
- Protease inhibitor mini tablet (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: A32955 )
- Sucrose (VWR, catalog number: 97063-788 )
- HEPES (Fisher Scientific, catalog number: BP310-500 )
- Hypotonic protease inhibitor solution (see Recipes)
- Sucrose buffer (see Recipes)
Equipment
- One 1 ml pipettor (Gilson, catalog number: F123602 )
- One pipet controller (VWR, catalog number: 613-4442 )
- One pair of scissors
- A set of stand (a stand and a clamp)
- Incubator
- Biosafety cabinet (Class II, Type A2)
- Centrifuge (Beckman Coulter, model: Allergra® X-22R ; Rotor: Beckman Coulter, model: SX4250 )
- Ultracentrifuge (Beckman Coulter, model: L-60 )
- Swing bucket rotor (Beckman Coulter, model: SW 28 )
- Fixed angle rotor (Beckman Coulter, model: 70 Ti )
- Belly dancer or orbital shaker
- Nitrogen cavitation chamber/vessel(Parr Instrument Company, catalog number: 4639 )
- Variable-Flow Peristaltic Pumps (Fisherbrand, Thermo Fisher)
Procedure
- Take two flasks (250 ml, 75 cm2) of confluent HEK293T cells (about 12 million cells per flask) that are expressing a fluorescently labeled membrane protein.
- Aspirate the media from the cells and rinse the cells with 5 ml 1x PBS buffer.
- Add 5 ml Versene solution and incubate at 37 °C for 5 min to detach the cells from the flasks.
- Collect the cell slurry in a 15 ml centrifuge tube.
- Centrifuge the slurry at 300 x g for 5 min at room temperature.
- Discard the supernatant (Versene solution) from the tube by careful aspiration and add 3 ml hypotonic protease inhibitor solution (Recipe 1) in the centrifuge tube. Resuspend the cells using pipetting in and out or swirling.
- Place the tube in an ice bucket and shake the bucket on an orbital shaker for 10 min to facilitate swelling of the cells.
- Transfer the cell slurry from the centrifuge tube into a pre-chilled (on ice) nitrogen cavitation chamber/vessel. Attach the vessel head and flow in nitrogen gas up to 600 psi. Incubate the cavitation chamber under pressure on ice for 20 min. Slowly open the valve to simultaneously release the pressure and collect the cell lysate into a 15 ml centrifuge tube (Figure 1). Keep the tube in ice until the cell lysate is added on top of the OptiPrep gradient.
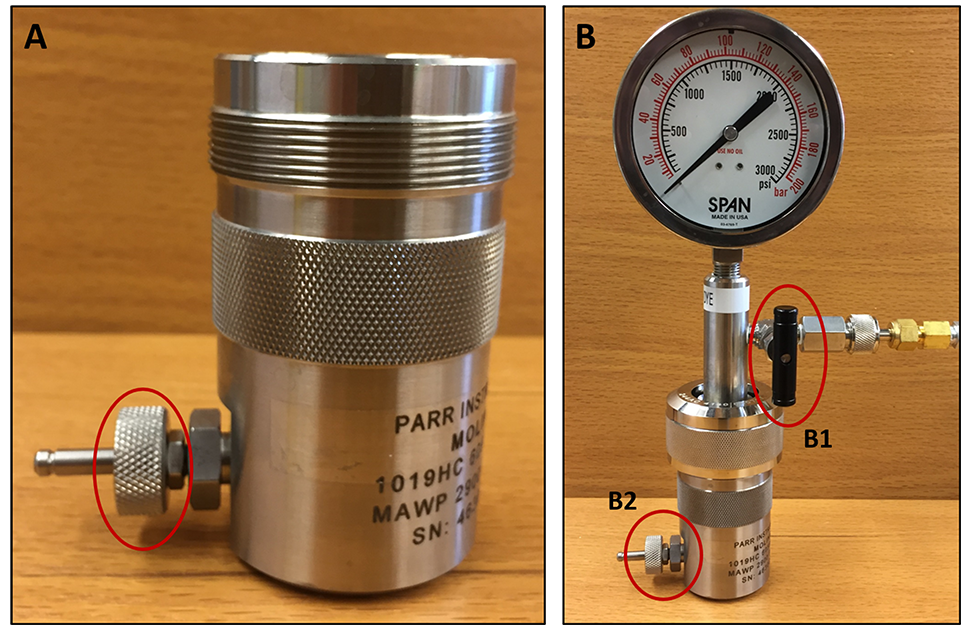
Figure 1. Instrumental setup used for nitrogen cavitation. A. Take the precooled nitrogen cavitation chamber, close the valve (red circle) and add the cell slurry into the chamber. B. Attach the vessel head and open valve B1 to flow nitrogen gas inside the chamber and close valve B1 once the pressure reaches 600 psi. Place this chamber into ice for 5 min. Take the chamber out from the ice and slowly open valve B2 to simultaneously release the pressure and to collect the cell lysate into a 15 ml centrifuge tube. - Make the OptiPrep gradient in an Ultra-Clear ultracentrifuge tube as follows:
- Cut a wine cork into ~2 mm thick circular slice using a razor blade (Figure 2A). Cut along the perimeter of the cork slice using a pair of scissors so that it can easily fit into the Ultra-clear ultracentrifuge tube. Wash the cork slice with water to remove debris. Leave it in sterile water until adding it to the ultracentrifuge tube.
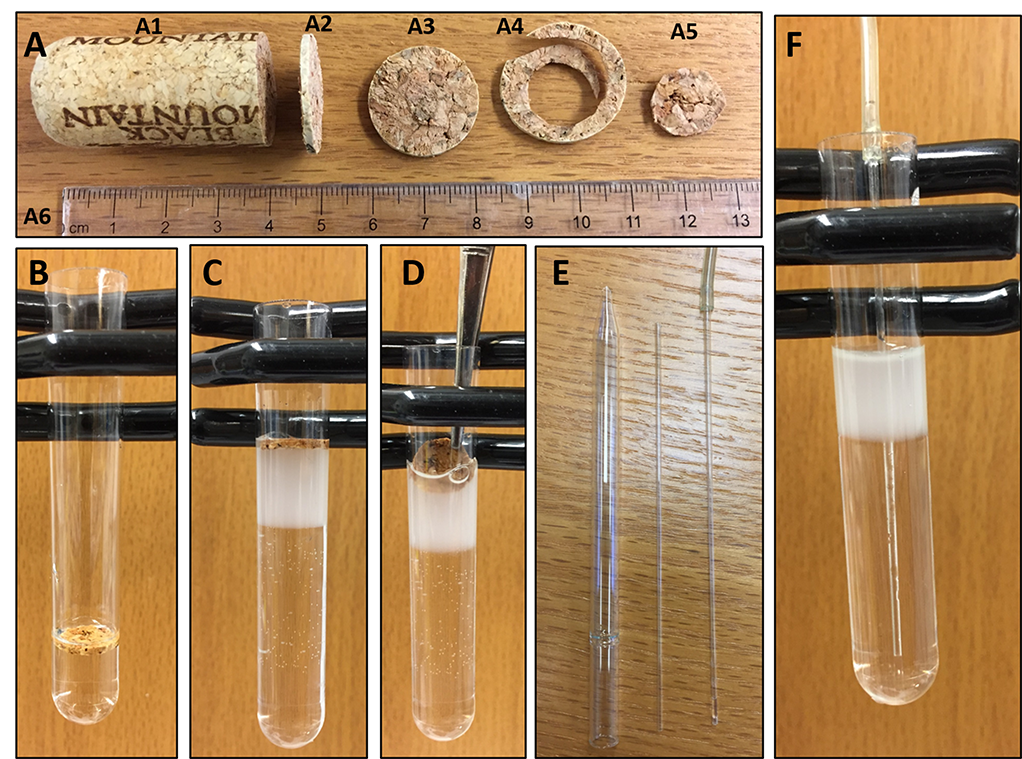
Figure 2. Pictorial presentation of the collection steps for ER and plasma membrane vesicles. A. A wine cork (A1) is cut into a slice of about 2 mm thickness (A2). This slice (A3) is cut along the circumference to discard the outer area (A4) and keep a small circular slice (A5) which can easily fit into an Ultra-Clear ultracentrifuge tube. A scale (A6) is placed on the image for comparison of sizes. B. The small slice of cork is inserted into the tube and floats on the solution. C. As additional solution (20%, and 10% OptiPrep) is added the cork rises to the top. After the addition of all layers, the cork floats atop the solution. D. The cork slice is removed using a forceps without disturbing the gradient. E. A Pasteur pipette is broken to obtain the narrow tube which is inserted in the inlet tubing of a peristaltic pump. F. The resultant inlet tubing of the pump is inserted at the bottom-center of the Ultra-Clear tube containing the gradient solution. - Prepare 30%, 20%, and 10% OptiPrep solutions by diluting the original 60% OptiPrep solution with sucrose buffer (Recipe 2) into three 50 ml tubes. Calculations associated with the preparation of 30 ml of each solution can be found in Table 1.
Table 1. Preparation of 30, 20 and 10% OptiPrep solution from 60% OptiPrep solution with sucrose buffer. The original OptiPrep solution obtained from the supplier contains 60% OptiPrep.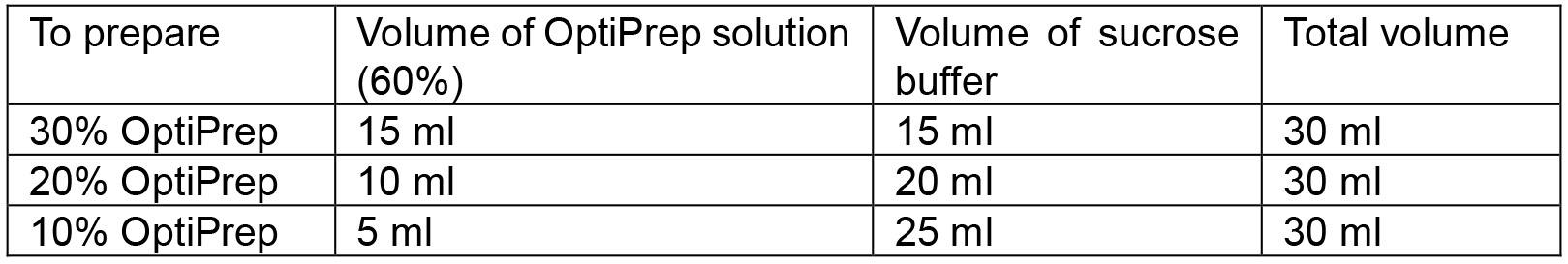
- In an Ultra-Clear ultracentrifuge tube, using a 5-ml Serological pipette and a pipette controller, add 3 ml 30% OptiPrep solution, then add the cork slice on top of the solution using a forceps (Figure 2B). Now add dropwise 3 ml 20% OptiPrep, then 3 ml 10% OptiPrep and finally 3 ml cell lysate on top of the cork slice. With the dropwise addition of solution in the tube, the cork slice stays floating on top of the solution and thus helps to avoid mixing of the added solution with the solution already present in the tube (Figure 2C).
Note: Three distinct interfaces should be visible indicating 4 layers of solutions of different densities. The interfaces are clearly visible when the tube is placed against a light source. - Carefully remove the wine cork using a forceps without disturbing the gradient (Figure 2D).
- Cut a wine cork into ~2 mm thick circular slice using a razor blade (Figure 2A). Cut along the perimeter of the cork slice using a pair of scissors so that it can easily fit into the Ultra-clear ultracentrifuge tube. Wash the cork slice with water to remove debris. Leave it in sterile water until adding it to the ultracentrifuge tube.
- Centrifuge the ultracentrifuge tube containing OptiPrep gradient in a swing bucket rotor at 25,000 rpm (112,000 x g) for 90 min at 4 °C.
- After the centrifugation, three layers of vesicles should be observable: between cell lysate and 10% OptiPrep, 10% and 20% OptiPrep, and 20% and 30% OptiPrep (as shown in Figure 3).
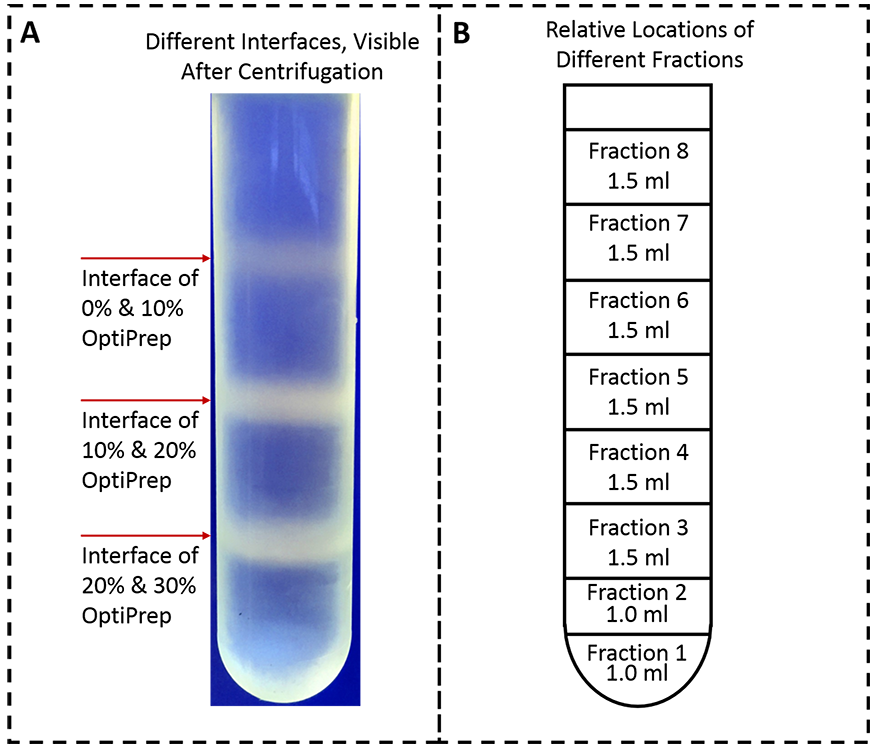
Figure 3. Presence of vesicles between the interfaces of OptiPrep solutions and the relative positions of different fractions collected. A. Three layers of vesicles are visible: on the interface of 0% and 10% OptiPrep, 10% and 20% OptiPrep and 20% and 30% OptiPrep. This image was taken with a mobile phone camera. B. By inserting the inlet tubing of a peristaltic pump in the bottom-center of the tube, collect fractions 1 and 2 in series, 1.0-ml each, then fractions 3-8 in a series, 1.5 ml each. - Use a peristaltic pump to collect ER and PM fractions as follows:
- Place the Ultra-Clear centrifuge tube vertically using a stand and a clamp.
- Break the glass Pasteur pipette at the junction of the small-diameter tube and large diameter tube and insert the small-diameter tube inside the inlet tubing of the peristaltic pump (Figure 2E).
- Carefully, insert the inlet tube of the peristaltic pump in the ultracentrifuge tube (fixed with a clamp and stand) until it touches the bottom-center of the centrifuge tube (Figure 2F). It is necessary to make sure that the gradient is not disturbed by the insertion process.
- Start the peristaltic pump to collect different fractions keeping the interfaces in the same fraction (Figure 3B). To do so, collect fractions 1 and 2, 1 ml each and fractions 3 to 8, 1.5 ml each.
- According to our Western blot results with anti-calnexin as an ER marker and anti-sodium potassium ATPase as a plasma membrane marker, fractions 3 and 4 contain only the ER and fractions 7 and 8 include only the plasma membrane vesicles (Figure 4). However, we have selected fractions 3 and 7 as the ER and plasma membrane vesicles respectively for our experiments.
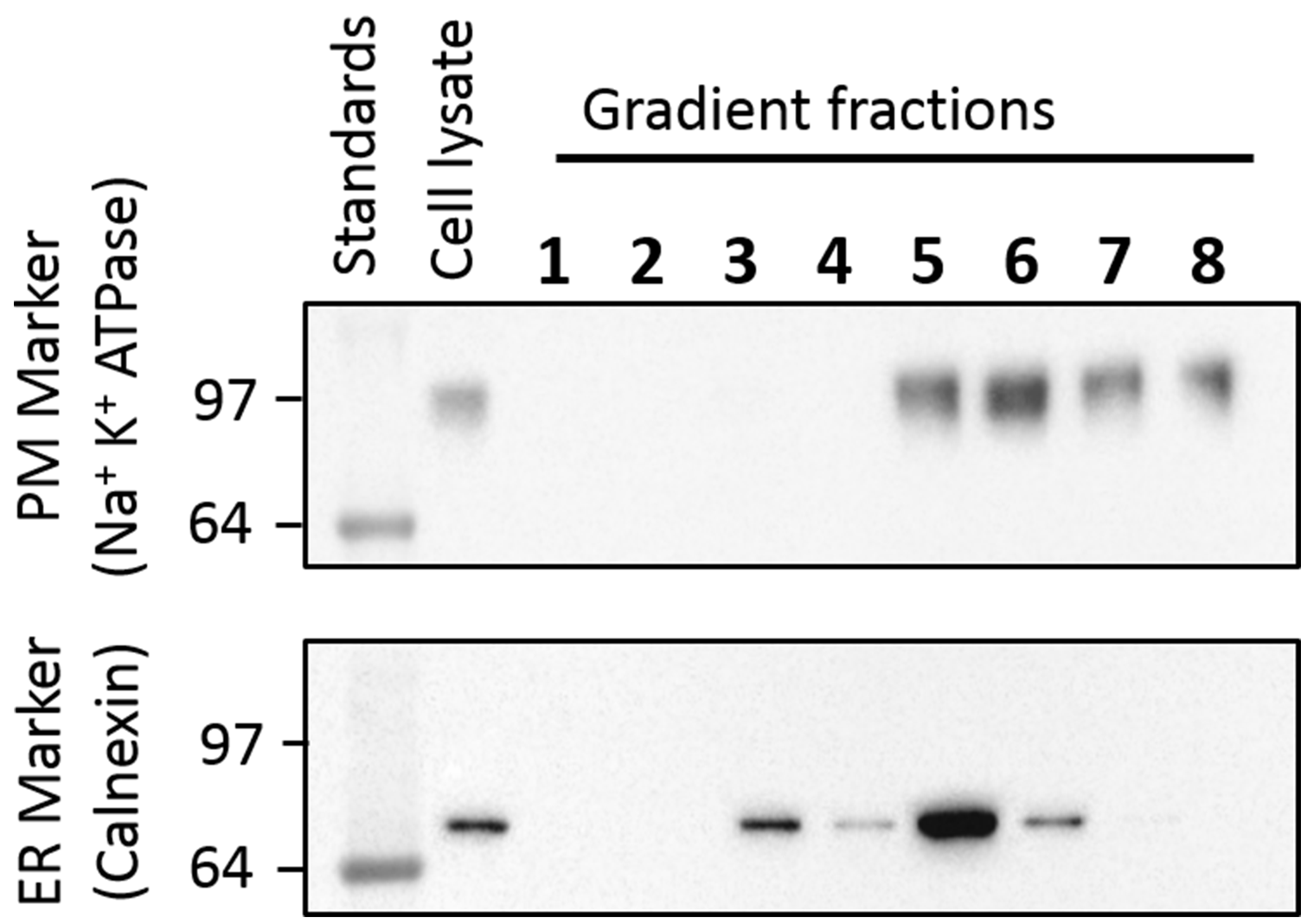
Figure 4. Western blot of the different fractions collected from OptiPrep gradient to isolate single receptors into the ER and plasma membrane specific vesicles. Anti-sodium potassium ATPase antibody (1:200 dilution) was used to bind with the plasma membrane marker, sodium potassium ATPase, in the OptiPrep gradient (upper panel). An anti-calnexin antibody (1:200 dilution) was employed to bind with the ER marker, calnexin, in the gradients (lower panel). An HRP conjugated mouse anti-Rabbit secondary antibody (1:5,000 dilution) and ClarityTM Western ECL Substrate on a ChemiDoc imaging system (Bio-Rad) were utilized to locate the ER and Plasma membrane markers on the gels. The plasma membrane marker was found in the cell lysate and fractions 5 to 8 and the ER marker was located in the cell lysate and fractions 3 to 6. Therefore, fractions 3 and 4 contain only the ER vesicles; fractions 7 and 8 contain only the plasma membrane vesicles. We selected fraction 3 for ER vesicles and fraction 7 for the plasma membrane vesicles. - Place the Ultra-Clear centrifuge tube vertically using a stand and a clamp.
- Dilute the ER and the plasma membrane fractions with 1x PBS buffer to 1:3 and centrifuge at 30,000 rpm (100,000 x g) for 1 h at 4 °C with the fixed angle rotor.
- Resuspend the pellets each in 600 μl 1x PBS buffer or sucrose buffer to obtain ER and plasma membrane derived vesicles. We usually aliquot the vesicle solution into two or three 1.5-ml tubes and store at -80 °C. We use the vesicles within 7 days.
Note: If a fraction contains no vesicles or a very small amount of vesicles, there will be no visible pellet on the surface of the tube after centrifugation. In this case, we collected the sample from the area where the vesicles might have present. It can be noted that a pellet is formed on the surface of the tube furthest from the center of the rotor and the possible location of the pellet can easily be assessed based on the location of the visible pellets in the other tubes.
At this point, a fraction of the ER and plasma membrane derived vesicles will contain the protein of interest. For single molecule fluorescence studies, it is necessary to start with cells expressing the target protein conjugated with a fluorescent protein. Single receptor containing vesicles can be employed to study the stoichiometric assembly of the receptors by step-wise photobleaching technique, to probe ligand-receptor interaction using fluorescence correlation spectroscopy and to understand dynamics and conformational changes of the receptor using single molecule Forster resonance energy transfer (smFRET).
Recipes
- Hypotonic protease inhibitor solution
10 mM Tris-HCl
10 mM NaCl
1.5 mM MgCl2
0.2 mM CaCl2
Adjust pH to 7.4
One protease inhibitor tablet per 10 ml buffer - Sucrose buffer
250 mM sucrose
10 mM HEPES
Adjust pH to 7.4
Acknowledgments
This work was supported in part by the NIDA training grant DA016176 (AMF) and NIDA DA038817. This work was adapted from Fox-Loe et al., 2017, J Biol Chem 292(51): 21159-21169. We declare no conflict of interest or competitive interest related to this publication.
References
- Andersson, H., Baechi, T., Hoechl, M. and Richter, C. (1998). Autofluorescence of living cells. J Microsc 191(Pt 1): 1-7.
- Grady, S. R., Drenan, R. M., Breining, S. R., Yohannes, D., Wageman, C. R., Fedorov, N. B., McKinney, S., Whiteaker, P., Bencherif, M., Lester, H. A. and Marks, M. J. (2010). Structural differences determine the relative selectivity of nicotinic compounds for native α4β2*-, α6β2*-, α3β4*- and α7-nicotine acetylcholine receptors. Neuropharmacology 58(7): 1054-1066.
- Graham, J., Ford, T. and Rickwood, D. (1994). The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal Biochem 220(2): 367-373.
- Fox, A. M., Moonschi, F. H. and Richards, C. I. (2015). The nicotine metabolite, cotinine, alters the assembly and trafficking of a subset of nicotinic acetylcholine receptors. J Biol Chem 290(40): 24403-24412.
- Fox-Loe, A. M., Moonschi, F. H. and Richards, C. I. (2017). Organelle-specific single-molecule imaging of α4β2 nicotinic receptors reveals the effect of nicotine on receptor assembly and cell-surface trafficking. J Biol Chem 292(51): 21159-21169.
- Henderson, B. J. and Lester, H. A. (2015). Inside-out neuropharmacology of nicotinic drugs. Neuropharmacology 96(Pt B): 178-193.
- Lester, H. A., Xiao, C., Srinivasan, R., Son, C. D., Miwa, J., Pantoja, R., Banghart, M. R., Dougherty, D. A., Goate, A. M. and Wang, J. C. (2009). Nicotine is a selective pharmacological chaperone of acetylcholine receptor number and stoichiometry. Implications for drug discovery. AAPS J 11(1): 167-177.
- Lippincott-Schwartz, J., Presley, J. F., Zaal, K. J., Hirschberg, K., Miller, C. D. and Ellenberg, J. (1999). Chapter 16: Monitoring the dynamics and mobility of membrane proteins tagged with green fluorescent protein. In: Sullivan, K. F. and Kay, S. A. (Eds.). Methods in Cell Biology. Academic Press 261-281.
- Moonschi, F. H., Effinger, A. K., Zhang, X., Martin, W. E., Fox, A. M., Heidary, D. K., DeRouchey, J. E. and Richards, C. I. (2015). Cell-derived vesicles for single-molecule imaging of membrane proteins. Angew Chem Int Ed Engl 54(2): 481-484.
- Richards, C. I., Luong, K., Srinivasan, R., Turner, S. W., Dougherty, D. A., Korlach, J. and Lester, H. A. (2012). Live-cell imaging of single receptor composition using zero-mode waveguide nanostructures. Nano Lett 12(7): 3690-3694.
- Tan, Y. W., Hanson, J. A., Chu, J. W. and Yang, H. (2014). Confocal single-molecule FRET for protein conformational dynamics. Methods Mol Biol 1084: 51-62
- Ulbrich, M. H. and Isacoff, E. Y. (2007). Subunit counting in membrane-bound proteins. Nat Methods 4(4): 319-321.
- Wang, S., Vafabakhsh, R., Borschel, W. F., Ha, T. and Nichols, C. G. (2016) Structural dynamics of potassium-channel gating revealed by single-molecule FRET. Nat Struct Mol Biol 23: 31
- Walter, N. G. and Bustamante, C. (2014) Introduction to single molecule imaging and mechanics: seeing and touching molecules one at a time. Chem Rev 114(6): 3069-71.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Moonschi, F. H., Fox-Loe, A. M., Fu, X. and Richards, C. I. (2018). Mammalian Cell-derived Vesicles for the Isolation of Organelle Specific Transmembrane Proteins to Conduct Single Molecule Studies. Bio-protocol 8(9): e2825. DOI: 10.21769/BioProtoc.2825.
Category
Molecular Biology > Protein > Isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










