- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In-vitro and in-planta Botrytis cinerea Inoculation Assays for Tomato
(*contributed equally to this work) Published: Vol 8, Iss 8, Apr 20, 2018 DOI: 10.21769/BioProtoc.2810 Views: 14055
Reviewed by: Tuan Minh TranAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries
Piao Yang [...] Ye Xia
Oct 20, 2023 2808 Views
Abstract
Botrytis cinerea (B. cinerea) attacks many crops of economic importance, represents one of the most extensively studied necrotrophic pathogens. Inoculation of B. cinerea and phenotypic analysis of plant resistance are key procedures to investigate the mechanism of plant immunity. Here we describe a protocol for B. cinerea inoculation on medium and planta based on our study using the tomato-B. cinerea system.
Keywords: Botrytis cinereaBackground
B. cinerea causes serious loss in more than 200 crops worldwide, including many important vegetables and small fruit crops. The broad-spectrum pathogen can infect plant stem, leaf, flower and fruit to produce spores (Dean et al., 2012; van Kan et al., 2017), which prefer to occur under high humidity (Elad et al., 2007). The produced spores pose long lasting threat to diverse hosts (Elad et al., 2007). Based on its scientific and economic importance, B. cinerea was ranked as the second most important plant-pathogenic fungus (Dean et al., 2012). Among B. cinerea host plants, tomato (Solanum lycopersicum), an economically valuable species, also serves as a classic model to study plant immunity (Ryan, 2000; Sun et al., 2011; Rosli and Martin, 2015). To investigate the molecular basis of plant immunity to B. cinerea, we employ a routine procedure to produce B. cinerea spores on artificial media. In addition, we provide detailed methods to infect tomato plants or detached leaves with a controlled strength using the collected spores and quantify disease development. This protocol has been successfully used to reveal the transcriptional regulation of master regulator MYC2 in Jasmonate-mediated plant immunity (Du et al., 2017).
Materials and Reagents
- General lab materials, including:
Nylon mesh (Solarbio, catalog number: YA0964 )
Petri dish (9 cm)
Square Petri dish (10 x 10 cm)
Funnel (Conventional type, upper diameter: 8 cm)
1.5 ml microcentrifuge tube (USA Scientific, catalog number: 4036-3204)
Manufacturer: Eppendorf, catalog number: 022363204 .
15 ml centrifuge tube (Corning, catalog number: 430790 )
50 ml centrifuge tube (Shanghai Kirgen, catalog number: KG2821 ) - Pipette tips
- Micropore tape (3M, MicroporeTM, catalog number: 1530S-1 )
- Nutritional soil (Miracle-Gro® Garden Soil for Vegetables with total N 0.68%, P2O5 0.27% and K2O 0.36%)
- Tomato (Solanum lycopersicum) cv M82
- B. cinerea B05.10
- V8 juice (Campbell, 100%, original vegetable juice)
- Calcium carbonate (CaCO3) (Sigma-Aldrich, catalog number: V900138 )
- Bacto-agar (BD, BactoTM, catalog number: 214010 )
- Mycological peptone (Sigma-Aldrich, catalog number: 77199 )
- Sodium phosphate (Sigma-Aldrich, catalog number: 342483 )
- Maltose (Sigma-Aldrich, catalog number: M5885 )
- 2x V8 agar medium (see Recipes)
- SMB medium (see Recipes)
- 0.8% agar medium (see Recipes)
Equipment
- Pipettes (Gilson, Pipetman® G)
- Water bath (YIHENG, model: DK-80 )
- Autoclave (Panasonic Healthcare, model: MLS-3781L )
- Customized transparent plastic box (materials: Polymeric Methyl Methacrylate, L/W/H: 50/50/50 centimeter, open at the top, Figure 1A) or any incubators that can be used alternatively
- Centrifuge (Eppendorf, models: 5810 R and 5424 )
- Microscope (ZEISS, model: Axio Imager Z2 ; 20x objective plan-APOCHROMAT; 10x objective EC plan-NEOFLUAR) or other light microscopes
- Counting chamber (QIUJING® KB-K-25, 0.1 mm, 1/400 mm2)
Software
- ImageJ (https://imagej.nih.gov/ij/download.html)
- Microsoft Excel
Procedure
- Preparation of seedling plants for infection
- Seeds treatment and germination
- Put tomato seeds into a cheesecloth or nylon bag and then incubate in a 50 °C water bath for 25 min. Briefly submerge the seeds in tap water to cool down to room temperature.
After that, transfer the seeds to a 10% trisodium phosphate (TSP) solution for 15 min and rinse (at least 5 times) in autoclaved distilled water for 5 min at room temperature to remove residual disinfectant. - Germinate tomato seeds for 48 h on a wet filter paper at room temperature in the dark (Figure 1B). Sow the germinated seeds in soil and cultivate in growth chambers under cycles of 16 h light at 25 °C and 8 h darkness at 18 °C. (The light intensity for light cycle was 200 μM photons m-2 sec-1 in our growth chamber.) For disease assay, the tomato seedlings are grown for 4 weeks (Figure 1C).
Note: In our lab, we use Miracle-Gro® Garden Soil (for Vegetables) for plant incubation, which is pre-sterile and less labor cost. Because plants grow faster in this soil, thus we use 4-week plants for the disease assay.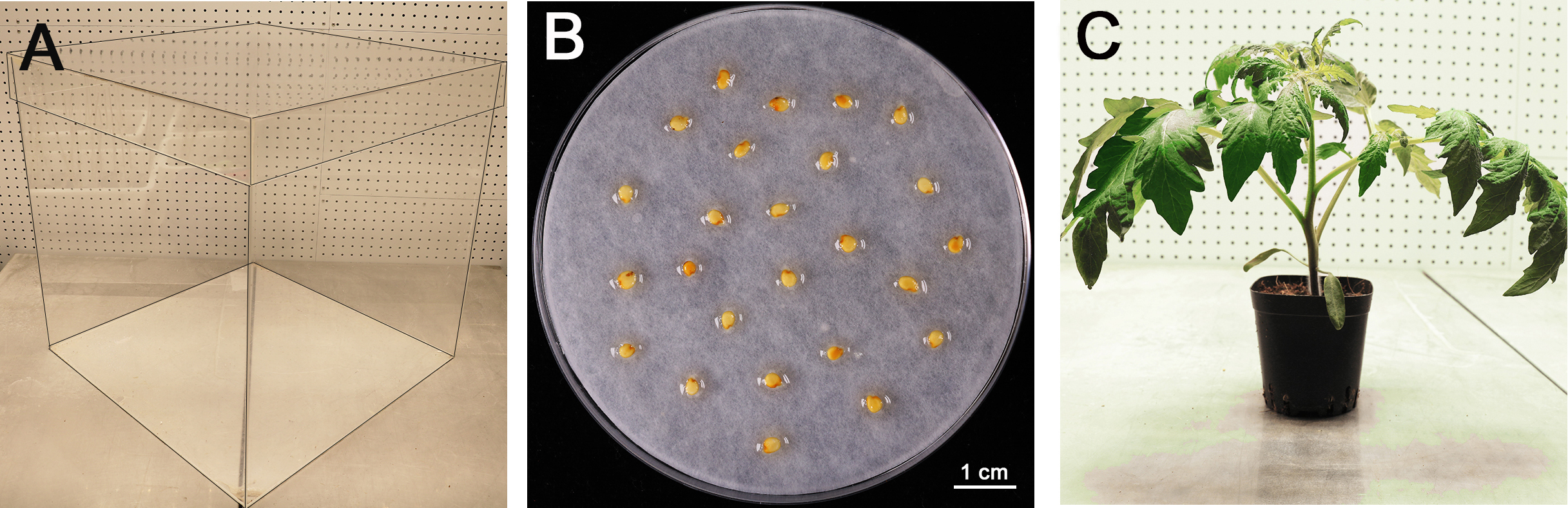
Figure 1. Preparation of tomato seedlings for infection. A. Customized transparent plastic box (edge highlighted with black lines) for plant inoculation, with 50 x 50 x 50 centimeter (cm). B. Tomato seeds on wet filter paper for germination after treatments. Scale bar = 1 cm. C. A 4-week-old tomato plant for inoculation assay.
- Put tomato seeds into a cheesecloth or nylon bag and then incubate in a 50 °C water bath for 25 min. Briefly submerge the seeds in tap water to cool down to room temperature.
- Seeds treatment and germination
- Fungal strains and growth conditions
- B. cinerea B05.10 used in this study is routinely maintained on 2x V8 agar for 21 days at 20 °C under a 12-h photoperiod prior to spore collection (Figure 2A).
- Harvest spores by scrubbing the plates with pipette tips until the tips have about 2 ml amount of tissues, then suspend in 5 ml 1% SMB. Filter the suspensions through nylon mesh on a funnel to remove mycelia (Figure 2B, Video 1). Video 1. How to collect spores
- Measure the concentration of spores with a counting chamber. Figure 2B shows the small squares of counting chamber we used under 20x objective. The counting chamber is 0.1 mm sample depth. The small square is 1/400 mm2 as shown in Figure 2B. It makes the volume of each small square 2.5 x 10-7 ml/square. Each spore in a small square is equal to 4 x 106 spores/ml. Adjust the concentration to 1 x 106 spores/ml for inoculation.
- Set a solution of 1% SMB without spores as the control treatment.
- B. cinerea B05.10 used in this study is routinely maintained on 2x V8 agar for 21 days at 20 °C under a 12-h photoperiod prior to spore collection (Figure 2A).
- Plant infection assay
The 4-week-old plants are selected for infection assay (Figure 1C).
For the pathogenicity test on detached leaves:- Individually harvest the leaflet of the third true leaves of 4-week-old plants and gently place in Petri dishes containing 25 ml 0.8% agar, with the petiole embedded in the medium (Video 1).
- Inoculate each leaflet with a single 5 μl spore suspensions droplet on the right or left of main midrib (Video 2). Cover plates with lids, followed by sealing with Micropore tape.Video 2. How to place a detached leaflet on medium
- Place the plates under the same conditions as for plant growth (Figure 2C). Monitor the disease development by scanning the plates 3 days after infection. The size of the infected area was measured with ImageJ.
For the pathogenicity test on living plants- Place 4-week-old plants in a transparent box (Figure 1A), which keeps the humidity at 100%. Inoculate the third true leaves with B. cinerea. Each leaflet is infected with a single 5 μl spore suspensions droplet on right or left of the main midrib (Video 3). Cover the box with lid and keep it under the same conditions used for plant growth (Figure 2D, Video 3).Video 3. Infecting a leaflet with a droplet of spore suspension
- Harvest the infected leaflets at different time points (0, 3, 6, 9, 12, 18, 24, 30, 36 h) after infection (HAI) for profiling resistance gene expression or at 3 days after infection (DAI) for monitoring disease development by imaging the leaflets.
- Then measure the size of the infected area on those leaflets with ImageJ (see Data analysis), following the procedure of the imageJ User Guide (https://imagej.nih.gov/ij/docs/index.html).
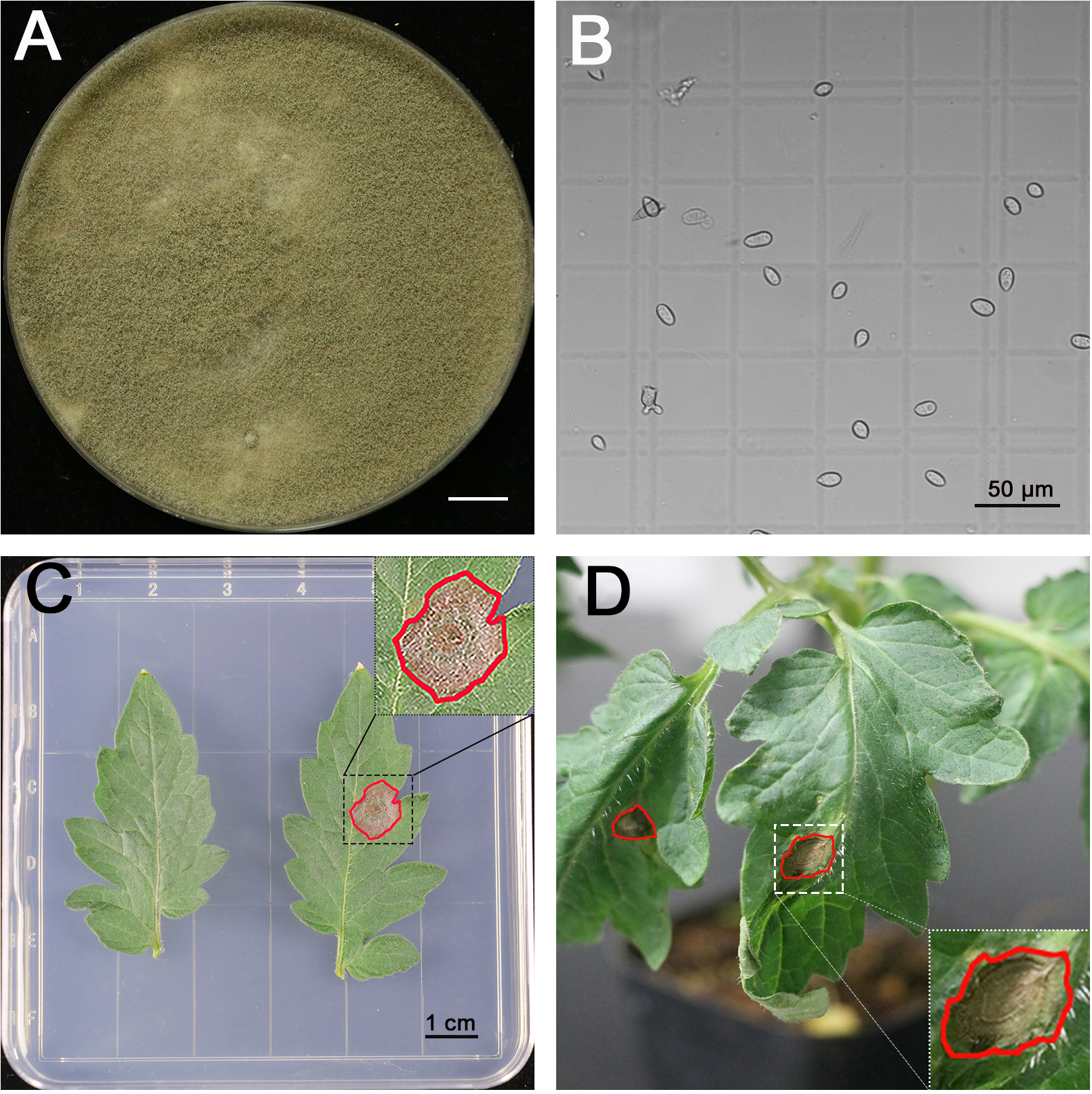
Figure 2. B. cinerea inoculation assay. A. 3-week-old B. cinerea strain on 2x V8 medium, scale bar for 1 cm. B. The spores of B. cinerea on counting chamber, scale bar for 50 µm. C. Pathogenicity test on detached leaves, the right leaflet was inoculated with a single 5 μl spore suspensions (106 spores/ml) droplet on the right or left of main midrib, while the left leaflet was set as control. The scale bar stands for 1 cm. D. Pathogenicity test on living plants, each leaflet was inoculated with a single 5 μl spore suspensions (106 spores/ml) droplet on the right or left of the main midrib. The areas infected by B. cinerea that developed for 3 days were highlighted with red circles.
- Individually harvest the leaflet of the third true leaves of 4-week-old plants and gently place in Petri dishes containing 25 ml 0.8% agar, with the petiole embedded in the medium (Video 1).
Data analysis
Images of the diseased leaflets were imported to software ImageJ. “Polygon selections” was used to select diseased area on the image. Then chose “measure” function from “Analyze” menu in software imageJ to calculate the lesion area. Data analysis was conducted in software Microsoft Excel.
Three independent experiments were conducted, each experiment included at least six replicates. All data from independent experiments were analyzed. The statistical test included average, STDEV and Student’s t-test calculation in excel. The t-test value of 0.05 is considered as significant difference and 0.01 for very significant difference. In the experiment, a droplet of spore suspension on leaf surface should develop to a diseased lesion with round shape. In some cases, the droplet spreads along with leaf vein and developed to an irregular shaped lesion, which should be avoided for analysis. Otherwise, all samples were collected for data analysis.
Notes
- Freshly produced spore should be used for the infection. The spore suspensions should be gently placed on leaf surface to prevent liquid spreading along with the tiny leaf vein.
- For detached leaf infection, the leaflets should cling to agar medium to maintain the moisture. After infection, the medium plates with infected leaves should be sealed with breathable tape to prevent the anaerobic respiration of leaves and keep moisture.
Recipes
- 2x V8 agar medium
360 ml V8 juice
2.00 g CaCO3
20 g Bacto-agar
Add ddH2O to 1,000 ml
Autoclave at 121 °C for 15 min - SMB medium
10.00 g Mycological peptone
40.00 g Maltose
Add ddH2O to 1,000 ml
Final pH (at 25 °C): 5.6 ± 0.2
Autoclave at 121 °C for 15 min - 0.8% agar medium
8.00 g Agar
Add ddH2O to 1,000 ml
Autoclave at 121 °C for 15 min
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFD0100603-10), by Tai-Shan Scholar Program from the Shandong Province. This protocol was adapted appropriately from previous work (Du et al., 2017). The authors declare that they have no competing interests.
References
- Elad, Y., Vivier, M. and Fillinger, S. (2016). Botrytis, the good, the bad and the ugly. In: Fillinger, S. and Elad, Y. (Eds). Botrytis: The fungus, the pathogen and its management in agricultural systems. Springer 1-15.
- Dahmen, H., Staub, Th. and Schwinn, F. J. (1982). Technique for long-term preservation of phytopathogenic fungi in liquid nitrogen. Phytopathol 73: 6.
- Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., Rudd, J. J., Dickman, M., Kahmann, R., Ellis, J. and Foster, G. D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13(4): 414-430.
- Du, M., Zhao, J., Tzeng, D. T. W., Liu, Y., Deng, L., Yang, T., Zhai, Q., Wu, F., Huang, Z., Zhou, M., Wang, Q., Chen, Q., Zhong, S., Li, C. B. and Li, C. (2017). MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29(8): 1883-1906.
- Rosli, H. G. and Martin, G. B. (2015). Functional genomics of tomato for the study of plant immunity. Brief Funct Genomics 14(4): 291-301.
- Ryan, C. A. (2000). The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta 1477(1-2): 112-121.
- Sun, J. Q., Jiang, H. L. and Li, C. Y. (2011). Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol Plant 4(4): 607-615.
- van Kan, J. A., Stassen, J. H., Mosbach, A., Van Der Lee, T. A., Faino, L., Farmer, A. D., Papasotiriou, D. G., Zhou, S., Seidl, M. F., Cottam, E., Edel, D., Hahn, M., Schwartz, D. C., Dietrich, R. A., Widdison, S. and Scalliet, G. (2017). A gapless genome sequence of the fungus Botrytis cinerea. Mol Plant Pathol 18(1): 75-89.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lian, J., Han, H., Zhao, J. and Li, C. (2018). In-vitro and in-planta Botrytis cinerea Inoculation Assays for Tomato. Bio-protocol 8(8): e2810. DOI: 10.21769/BioProtoc.2810.
Category
Plant Science > Plant immunity > Disease bioassay
Microbiology > Microbe-host interactions > In vitro model
Microbiology > Microbe-host interactions > Virus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link













