- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Bacterial Twitching Motility in Dense Colonies Using Transmitted Light Microscopy and Computational Image Analysis
Published: Vol 8, Iss 8, Apr 20, 2018 DOI: 10.21769/BioProtoc.2804 Views: 7745
Reviewed by: Andrea PuharTimo A LehtiAlexander B Westbye

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis
Mădălina Mocăniță [...] Vanessa M. D'Costa
Mar 5, 2024 2922 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2112 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1697 Views
Abstract
A method was developed to allow the quantification and mapping of relative bacterial twitching motility in dense samples, where tracking of individual bacteria was not feasible. In this approach, movies of bacterial films were acquired using differential interference contrast microscopy (DIC), and bacterial motility was then indirectly quantified by the degree to which the bacteria modulated the intensity of light in the field-of-view over time. This allowed the mapping of areas of relatively high and low motility within a single field-of-view, and comparison of the total distribution of motility between samples.
Keywords: BacteriaBackground
Pilus-mediated twitching motility represents a form of surface-associated bacterial movement that is independent of flagella. Twitching motility is utilized by many bacterial pathogens, including Neisseria gonorrhoeae and Pseudomonas aeruginosa, to interact with moist surfaces and translocate epithelial barriers. In P. aeruginosa, twitching motility is regulated by a large number of genes which allow both extension and retraction of type IV pili to effectively drag the bacterial cell across any given surface in response to environmental cues (Mattick, 2002; Whitchurch et al., 2004; Burrows, 2005). In our studies of P. aeruginosa pathogenesis, twitching motility contributes to bacterial exit from epithelial cells after internalization and bacterial traversal of multilayered corneal epithelia (Alarcon et al., 2009). In a murine model of corneal infection, twitching motility was important for P. aeruginosa virulence (Zolfaghar et al., 2003). Recently, we discovered that the glycoprotein DMBT1 found in mucosal fluids such as human tears and saliva was capable of inhibiting P. aeruginosa twitching motility (Li et al., 2017). In that study, we utilized a novel method to quickly and robustly quantify P. aeruginosa twitching motility. That protocol is presented herein.
The most direct way to quantify twitching motility would be to track all individual bacteria over time. This method was attempted as part of our original study. However, bacterial colonies have a complex 3D structure, with bacteria regularly traversing one another, making tracking feasible only near the colony edge, resulting in sampling bias. Previous methods for quantifying twitching motility also quantified motility only at the colony edge (Alarcon et al., 2009; Semmler et al., 1999). For our study, we wished to extend those methods to be able to quantify twitching behavior throughout a dense bacterial colony. Instead of focusing on the direction of motility, we focused on quantifying the degree of motility at any given point. This turned out to be a simpler problem to solve, since as bacteria move, they modulate light as they pass through a given point. By mapping out the relative magnitudes of this modulation over time, we were able to generate maps of regions of relatively high and low motility in dense, spatially complex colonies.
Materials and Reagents
- Glass slides (Fisher Scientific, Fisherbrand, catalog number: 12-550-15 )
- Cover slip (Fisher Scientific, Fisherbrand, catalog number: 12-545M )
- Small Petri dish (Corning, Falcon®, catalog number: 351029 )
- Large Petri dish (Sigma-Aldrich, catalog number: P5981-100EA)
Manufacturer: Excel Scientific, catalog number: D-902 . - Kimwipes (KCWW, Kimberly-Clark, catalog number: 34155 )
- Inoculation loop (Fisher Scientific, Fisherbrand, catalog number: 22-363-597 )
- Sterile wooden toothpick (any brand, autoclave at 121 °C for 15 min in a foil-covered container)
- Pseudomonas aeruginosa strain MPAO1 (Dr. Manoil Laboratory, University of Washington, Seattle, WA), or other piliated strain (or bacterial species) with functional twitching motility. (MPAO1 is a wild-type P. aeruginosa strain, and is available from the University of Washington, Seattle, WA. http://www.gs.washington.edu/labs/manoil/libraryindex.htm)
- Substance to be tested in the assay (e.g., human tear fluid)
- Deionized water
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Fisher Chemical, catalog number: M63-500 )
- Gellan gum (Alfa Aesar, catalog number: J63423 )
- Tryptone (BD, BactoTM, catalog number: 211705 )
- Sodium chloride (NaCl) (Fisher Scientific, Fisher Chemical, catalog number: S271-3 )
- Yeast extract (BD, BactoTM, catalog number: 288620 )
- Trypticase Soy Agar (TSA) powder (BD, DifcoTM, catalog number: 236950 )
- Twitching medium (see Recipes)
- Trypticase Soy Agar (TSA) plate (see Recipes)
Equipment
- Tweezers (Dumont, Dumoxel Nº5, Fine Science Tools, catalog number: 11252-30 )
- Bunsen burner
- 3.60 °C oven (Boekel Scientific, model: 133000 )
- Sterile Biosafety Cabinet (NUAIRE, model: NU-425-600 )
- Compound microscope with a high NA objective and digital camera (≥ 1.2). Our study used:
- Nikon Ti-E inverted wide-field fluorescence microscope (Nikon Instruments, model: Eclipse Ti-E )
- Uno-combined controller (Okolab) and stage-top incubation chamber to maintain samples at 37 °C (Okolab, model: H301-Nikon-TI-S-ER )
- CFI Plan Apo Lambda 60x NA 1.4 oil objective (Nikon Instrument, model: CFI Plan Apochromat Lambda (λ) Series )
- DS-Qi2 Monochrome CMOS Camera (Nikon Instrument, model: DS-Qi2 )
- Nikon Ti-E inverted wide-field fluorescence microscope (Nikon Instruments, model: Eclipse Ti-E )
- Computer. MacPro5.1 (2012), 2x 2.4 GHz 6-Core Intel Zeon, 64 GB 1333 MHz DDR 3 ECC
Software
- ImageJ (version 1.51n) or FIJI (http://fiji.sc/)
- R compiler (version x64 3.4.1) (https://www.r-project.org/)
Procedure
- Preparation of slides
- Sterilize each glass slide by holding it over a Bunsen flame with tweezers (also sterilized).
- Place slides (up to 4) in a large Petri dish.
- Quickly pour 1 ml of twitching medium (kept at ~60 °C) onto one surface of each slide and let it spread evenly (by gravity). The approximate thickness of the medium should be 1.5-2 mm.
- Place the slide(s) in a biosafety cabinet (BSL2) to cool and dry (20 min). Then, gently clean the underside of the slide with a dry Kimwipe to remove any residual twitching medium.
- Draw a circle (~5 mm diameter) on the backside of the slide to mark where to adsorb the substance to be tested (see Figure 1). Up to 3 circles can be drawn per slide.
- Drop 5 µl of the substance to be tested onto the medium overlying the circle (Figure 1). Make sure not to touch the medium. A 5 µl volume usually covers a ~5 mm circle.
- Dry the slide in a biosafety cabinet (BSL2) (15 min).
- If more samples are needed, Steps A6 and A7 can be repeated.
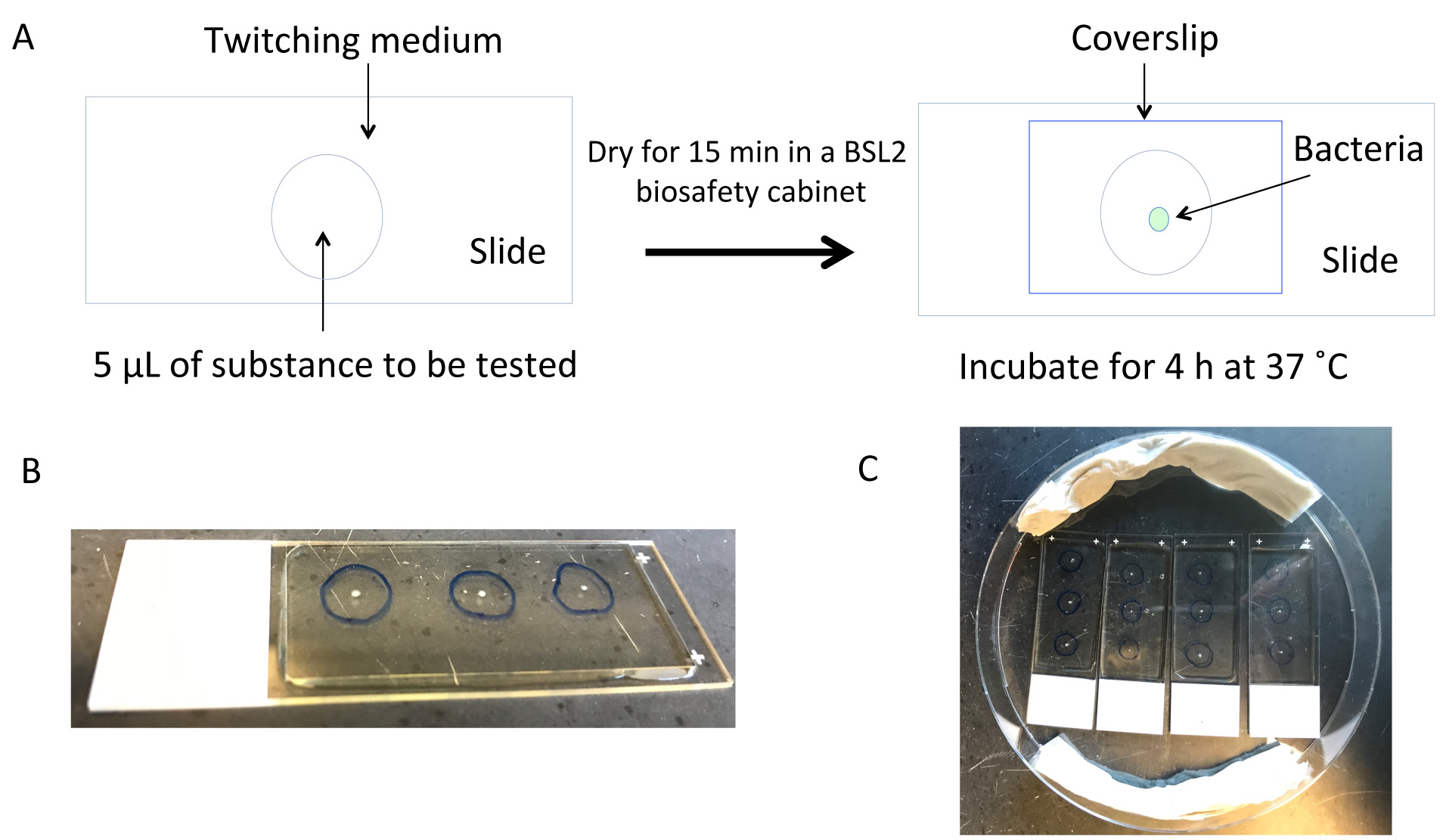
Figure 1. Experimental setup and images of the twitching assay. A. Schematic showing the experimental setup. Twitching medium (~60 °C) is poured onto a glass slide and allowed to cool and dry in a biosafety cabinet (BSL2) for 20 min. After adding the substance to be tested, the slide is dried again in the biosafety cabinet for 15 min before inoculation with bacteria. A coverslip is then placed over the inoculated substance to be tested, and the slide incubated for 4 h at 37 °C in a moist environment. B. Example of an inoculated slide with coverslip showing three circles, within each a replicate of the substance being tested. C. Example of a large Petri dish containing four inoculated slides with coverslips (e.g., a control slide versus three slides comparing different substances, or different concentrations of the same substance). Two wet paper towels are included to prevent drying of the twitching medium during the 4 h incubation. Although not shown, the Petri dish is covered with its lid during incubation to maintain moisture. - Sterilize each glass slide by holding it over a Bunsen flame with tweezers (also sterilized).
- Inoculate twitching medium
- From an overnight growth on TSA plates, scrape a few colonies with an inoculation loop and mix together on the plate. This helps to make sure that all inoculations originate from a homogeneous mix of bacteria, and not a single clone, reducing possible motility differences due to phenotypic variation. However, single colonies can be tested if suited to study goals.
- Transfer a very small amount of bacteria (~0.5 mm diameter) to the tip of a sterile toothpick.
- Use the toothpick to apply the bacteria lightly on the slide in the center of the circles, without piercing the twitching medium. A single, central point of inoculation is required.
- Use sterile tweezers to lay a coverslip over the medium and point of inoculation (don’t press down onto the inoculated point) (Figure 1).
- Wet a rolled paper towel with deionized water. Put in the large Petri dish with the slides to keep them moist (Figure 1). Cover the Petri dish.
- Incubate for 4 h at 37 °C.
- From an overnight growth on TSA plates, scrape a few colonies with an inoculation loop and mix together on the plate. This helps to make sure that all inoculations originate from a homogeneous mix of bacteria, and not a single clone, reducing possible motility differences due to phenotypic variation. However, single colonies can be tested if suited to study goals.
- Imaging
After the incubation in Step B6, acquire time-lapse movies of the bacteria at the edge of the point of inoculation (10 sec interval for 5 min duration) at 37 °C, using a 60x/1.4NA oil-immersion lens with differential interference contrast (DIC).
Data analysis
DIC movies of bacterial twitching motility were computationally analyzed using ImageJ. The code used for our analysis has been made available in a Github repository for reference (Smith, 2017). Specifically, the analysis normalizes the contrast across the field of view and between frames. This then allows for the indirect quantification of bacterial motility as the degree of light modulation over time, where regions of high motility will modulate light more than regions low motility. Maps of relative bacterial motility were created by taking the standard deviation of the intensity in the processed movies over time (see example, Figure 2). The total motility in each sample was then compared across experiments by plotting the distribution of the standard deviation as a notched box plot (see example, Figure 3), where two distributions are considered significantly different if the notches do not overlap.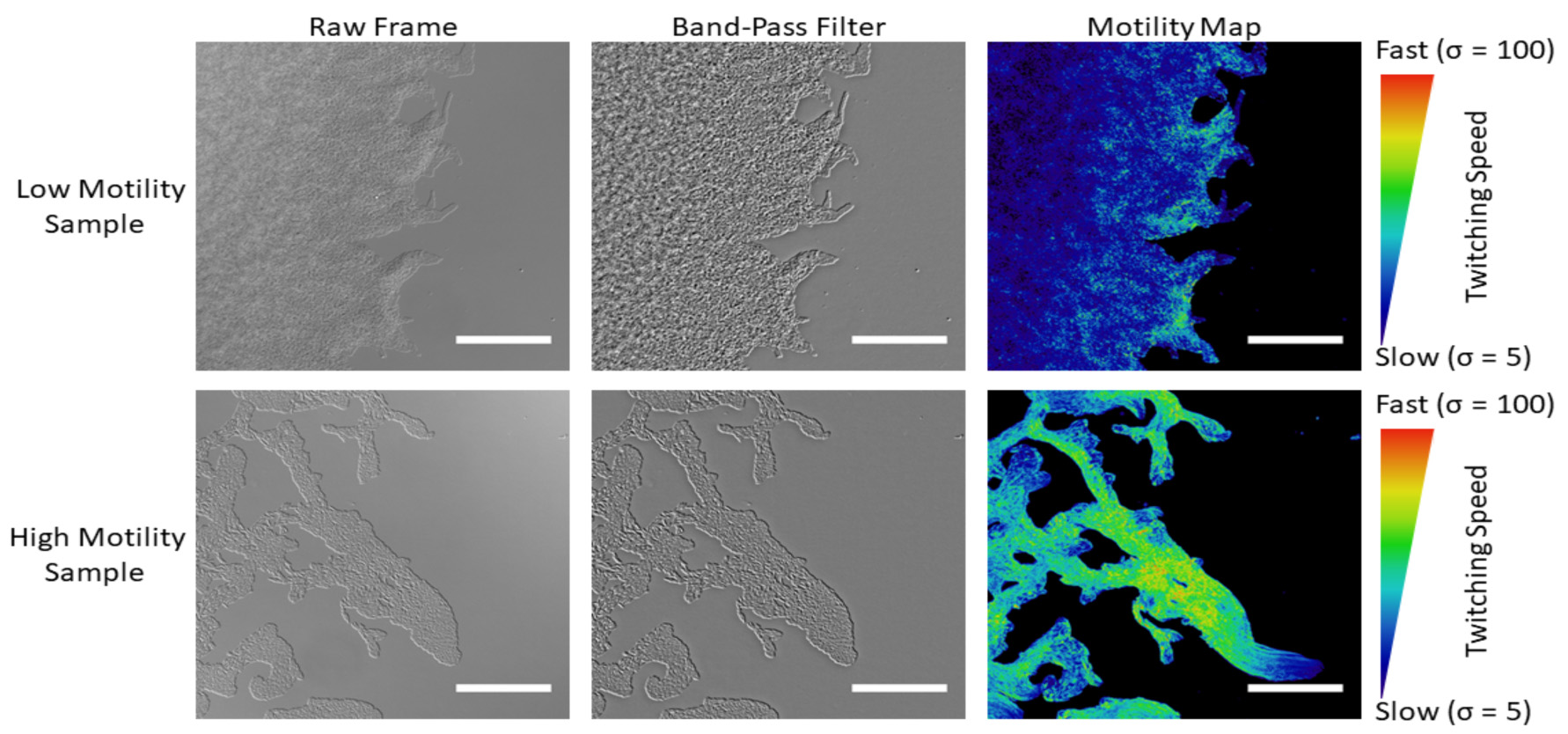
Figure 2. Creating motility maps from the raw movies. The raw movies are processed with a band-pass filter to normalize the contrast across the field of view, and to enhance the contrast specific to individual bacteria. A motility map is then generated by taking a standard deviation Z-projection of the processed movie. Scale bars = 50 μm.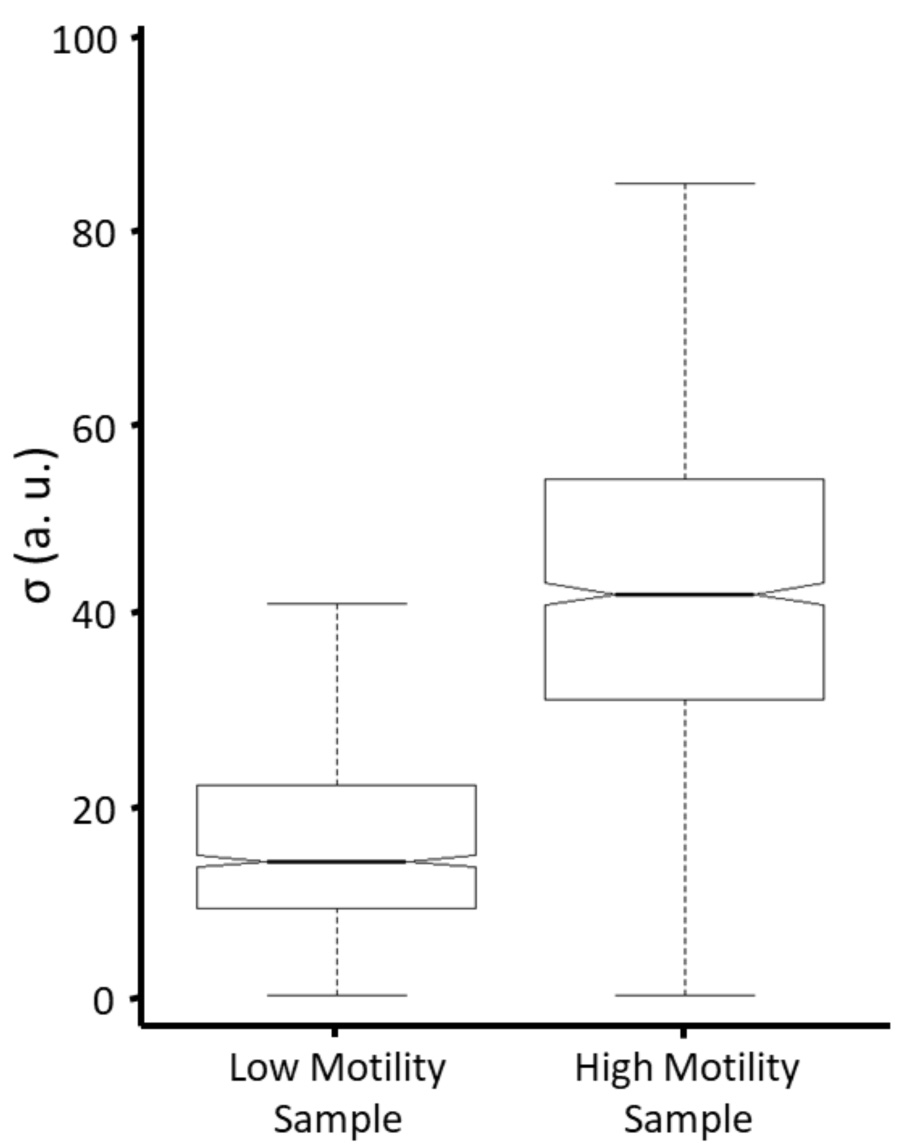
Figure 3. Comparing motility distributions between samples. The motility distributions between samples can be compared using notched box-plots, where the notches represent a 95% confidence interval for each distribution.
- Open the original raw movie in FIJI or ImageJ.
- Use a 2D median filter (Process → Filters → Median) to remove any hot pixels and other single pixel outliers from the movie (for our data, a median filter with a radius of 2.0 pixels worked best).
- Use a 2D band-pass filter (Process → FFT → Band-pass Filter) to normalize any vignetting and/or other low spatial frequency background non-uniformities, as well as remove high spatial frequency noise (for our data, we used a lower bound of 40 and an upper bound of 2). These numbers will need to be adjusted according to the sample and microscope, with the lower bound primarily affected by bacterial size (with smaller bacteria, e.g., Staphylococcus aureus possibly requiring a smaller lower bound), and the upper bound primarily affected by the ratio of the digital resolution/optical resolution, where the larger the ratio (i.e., where the camera resolution is much greater than the optical resolution) the larger the upper bound. The purpose of the band-pass is to remove any spatial frequencies that do not pertain to individual bacteria, therefore greatly suppressing any convolved modulations.
- Create a standard deviation Z-projection (Image → Stacks → Z project) of the movie to create a map of the magnitude of bacterial motility across the field of view.
- Generate a histogram of the standard deviation map (Analyze → Histogram) to get a distribution of bacterial motility within the field of view.
- Since the motility distribution may not be normal and instead heavily skewed, the distributions can be compared using notched boxplots (Figure 3). Notched boxplots are especially helpful when there are more than two distributions to compare in a set, as the notches allow for quick visualization of significant differences between all-possible pairwise combinations of samples (Krzywinski and Altman, 2014).
Notes
- For each substance tested, we typically used three replicates on a single slide. Each experiment would also include a twitching motility positive-control slide with three replicates. We did not observe a large difference in motility between replicate inoculations; however, if the colonies were too large, the colony edges could be difficult to discern.
- The noise from the camera and variation in the light source over time will also result in small modulations in intensity in the movie. Therefore, a minimum cutoff for the standard deviation needs to be chosen that completely excludes the background while not removing bacterial modulation. In our setup, we found that a cutoff of 5.0 worked best.
- To make the motility maps more clear, we used the ‘physics’ LUT provided with ImageJ (Image → Lookup Tables → Physics), which gives a spectral heat-map making differences in local motility more apparent. For added clarity, we set pixels with 0 intensity to black (Image → Color → Edit LUT).
- In order to use the scripts provided in the Github repository, first, go to the provided link and click on “Clone or download” and download the repository as a zip file. Unzip the files, then run the files in the following order:
- Drag and drop the file entitled “Measure stack standard deviation.ijm” into FIJI/ImageJ. This will load the code into the macro IDE. Then press “Run” in the IDE window. This will pull-up a user interface that will ask for the directory that contains all of your images to be analyzed, and then prompt you for the directory where the results should be saved. You will then be prompted to enter the lower and upper standard deviation cut-offs for the data. The code will automatically process the data and output a histogram for each sample into the output directory in a file called “Sample histogram.csv”.
NOTE: The current code will only process *.nd2 files; this extension can be changed on line 3 of the macro. - Open the file entitled “Boxplot code – R.r” in a text editor, and change the directory on line 8 to the directory that contains the histogram from (a). Then copy and paste the code into the R compiler and it will automatically create notched boxplots from the histograms.
Note: If the minimum and/or maximum standard deviation cut-offs were changed from the default values in the macro, they will have to be updated accordingly in line 5 in the R code. If you forgot the range, the bin values are stored in the output directory in a file called “Histogram bins.csv.”.
- Drag and drop the file entitled “Measure stack standard deviation.ijm” into FIJI/ImageJ. This will load the code into the macro IDE. Then press “Run” in the IDE window. This will pull-up a user interface that will ask for the directory that contains all of your images to be analyzed, and then prompt you for the directory where the results should be saved. You will then be prompted to enter the lower and upper standard deviation cut-offs for the data. The code will automatically process the data and output a histogram for each sample into the output directory in a file called “Sample histogram.csv”.
Recipes
- Twitching medium
- Dissolve 0.1 g MgSO4·7H2O, 0.8 g Gellan gum, 0.4 g tryptone, 0.2 g yeast extract and 0.2 g NaCl in ~80 ml of distilled H2O and shake until the solutes have dissolved
- Adjust the final volume of the solution to 100 ml with H2O
- Sterilize by autoclaving in the liquid cycle. Keep the medium at 60 °C until pouring to prevent solidification
- Dissolve 0.1 g MgSO4·7H2O, 0.8 g Gellan gum, 0.4 g tryptone, 0.2 g yeast extract and 0.2 g NaCl in ~80 ml of distilled H2O and shake until the solutes have dissolved
- Trypticase Soy Agar (TSA) plate
- Suspend 40 g of TSA powder in 1 L of purified water. Mix thoroughly and sterilize by autoclaving in the liquid cycle
- Cool to ~50 °C and pour approximately 20 ml into each small Petri dish on a level surface
- Cover the Petri dish with its lid and let solidify at room temperature (~2 h or until solid and dry)
Acknowledgments
This protocol was originally presented in Li et al., 2017, PLoS Pathogens. This work was supported by the National Institutes of Health; EY011221 (SMJF), EY024060 (SMJF) and EY003176. The authors have no conflicts of interest to declare. P. aeruginosa strain MPAO1 was obtained from the University of Washington (Dr. Manoil laboratory), Seattle, WA where it was previously used to generate the P. aeruginosa PAO1 transposon mutant library funded by NIH P30 DK089507.
References
- Alarcon, I., Evans, D. J. and Fleiszig, S. M. (2009). The role of twitching motility in Pseudomonas aeruginosa exit from and translocation of corneal epithelial cells. Invest Ophthalmol Vis Sci 50(5): 2237-2244.
- Burrows, L. L. (2005). Weapons of mass retraction. Mol Microbiol 57(4): 878-888.
- Krzywinski, M. and Altman, N. (2014). Visualizing samples with box plots. Nat Methods 11(2): 119-120.
- Li, J., Metruccio, M. M. E., Smith, B. E., Evans, D. J. and Fleiszig, S. M. J. (2017). Mucosal fluid glycoprotein DMBT1 suppresses twitching motility and virulence of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Pathog 13(5): e1006392.
- Mattick, J. S. (2002). Type IV pili and twitching motility. Annu Rev Microbiol 56: 289-314.
- Semmler, A. B., Whitchurch, C. B. and Mattick, J. S. (1999). A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145 (Pt 10): 2863-2873.
- Smith, B. E. (2017). Bacterial-Twitching-Quantification. Github repository. https://github.com/Llamero/Bacterial-Twitching-Quantification.
- Whitchurch, C. B., Leech, A. J., Young, M. D., Kennedy, D., Sargent, J. L., Bertrand, J. J., Semmler, A. B., Mellick, A. S., Martin, P. R., Alm, R. A., Hobbs, M., Beatson, S. A., Huang, B., Nguyen, L., Commolli, J. C., Engel, J. N., Darzins, A. and Mattick, J. S. (2004). Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52(3): 873-893.
- Zolfaghar, I., Evans, D. J. and Fleiszig, S. M. (2003). Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect Immun 71(9): 5389-5393.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Smith, B. E., Li, J., Metruccio, M., Wan, S., Evans, D. J. and Fleiszig, S. M. J. (2018). Quantification of Bacterial Twitching Motility in Dense Colonies Using Transmitted Light Microscopy and Computational Image Analysis. Bio-protocol 8(8): e2804. DOI: 10.21769/BioProtoc.2804.
Category
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell imaging > Live-cell imaging
Cell Biology > Cell movement > Cell motility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











