- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of RNA Mango Tagged Native RNA-protein Complexes from Cellular Extracts Using TO1-Desthiobiotin Fluorophore Ligand
Published: Vol 8, Iss 7, Apr 5, 2018 DOI: 10.21769/BioProtoc.2799 Views: 8767
Reviewed by: Gal HaimovichAnca SavulescuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Charging State Analysis of Transfer RNA from an α-proteobacterium
Liang Yin and Caroline S. Harwood
Dec 5, 2020 3507 Views

Ribosome Purification from an α-proteobacterium and rRNA Analysis by Northern Blot
Liang Yin and Caroline S. Harwood
Dec 5, 2020 3323 Views

Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies
Parthasarathy Manikandan [...] Mahavir Singh
Jul 5, 2023 2172 Views
Abstract
A native purification strategy using RNA Mango for RNA based purification of RNA-protein complexes is described. The RNA Mango aptamer is first genetically engineered into the RNA of interest. RNA Mango containing complexes obtained from cleared cellular native extracts are then immobilized onto TO1-Desthiobiotin saturated streptavidin agarose beads. The beads are washed to remove non-specific complexes and then the RNA Mango containing complexes are eluted by the addition of free biotin to the beads. Since the eluted complexes are native and fluorescent, a second purification step such as size exclusion chromatography can easily be added and the purified complexes tracked by monitoring fluorescence. The high purity native complexes resulting from this two-step purification strategy can be then used for further biochemical characterization.
Keywords: RNA MangoBackground
Current RNA tags suffer from limitations such as poor KD, large size, potential biological interference or lack of intrinsic fluorescence (Panchapakesan et al., 2015). RNA Mango is small, can be simply integrated into stem-loop structures, in particular, GNRA tetraloops, is biologically tolerated and above all has a high affinity for its thiazole orange-based (TO1) ligand, TO1-Desthiobiotin (TO1-Dtb). This allows Mango tagged complexes to be easily bound and washed on streptavidin beads (summarized in Figure 1). The Mango:TO1-Desthiobiotin complex is highly fluorescent and can be eluted from streptavidin beads by the addition of biotin. The fluorescence of Mango tagged native complexes allows additional purification steps to be used to obtain highly purified native complexes. 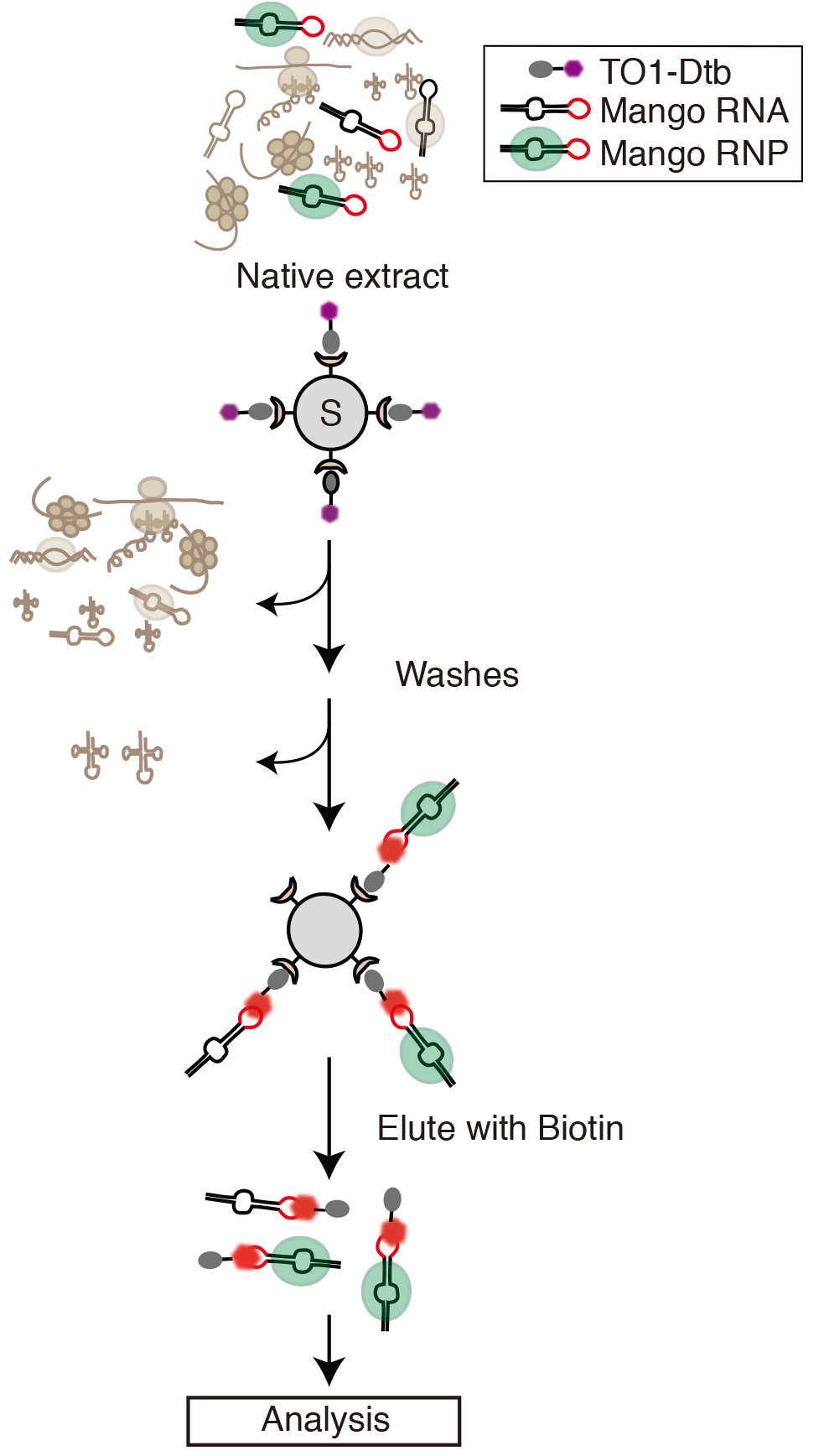
Figure 1. Purification of Mango tagged RNA-protein complexes out of native extract. RNA constructs are designed (Procedure A, Figure 2) so that the Mango tag is located in a biologically compatible location. After bacterial expression of this construct, a native extract containing the Mango tagged RNA (Mango tag shown in red highlight), is prepared (Procedure B). Mango tagged RNA and RNA complexes are then bound to streptavidin beads (Black circle containing S) that have been derivatized with TO1-Desthiobiotin (Dtb, Procedure C). The thiazole orange moiety of TO1-Dbt is shown in purple and becomes highly fluorescent once bound by Mango (bright red highlight). Bound complexes are then extensively washed in native conditions (Procedure D). After washing fluorescent Mango tagged complexes can be eluted in native conditions by addition of biotin (Procedure E). Downstream analysis including further purification steps can then be simply implemented (Procedures F and G).
Materials and Reagents
- Low retention pipette tips (Fisher Scientific, catalog numbers: 02-717-134 , 02-717-143 , 02-707-511 )
- Microcentrifuge tubes, pre-lubricated (Corning, Costar®, catalog numbers: 3206 , 3207 )
- 96-well black wall, clear bottom, non-binding, non-sterile, Greiner Bio-One microtiter plate (Greiner Bio One International, catalog number: 655906 )
- Tricorn XK 16/40 SEC column (GE Healthcare, catalog number: 28988938 )
- Bacterial cells (E. coli) with RNA Mango tagged RNA of interest
- Liquid nitrogen
- TO1-PEG3-Desthiobiotin (ABM, catalog number: G956 )
- High capacity streptavidin agarose beads (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 20357 )
- Superdex 200 resin (GE Healthcare, catalog number: 17104301 )
- SYBR Green II RNA Gel Stain (Thermo Fisher Scientific, InvitrogenTM, catalog number: S7564 )
- 19:1 Acrylamide:Bis 40% (Thermo Fisher Scientific, catalog number: AM9022 )
- N,N,N’,N’-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281 )
- Ammonium persulphate (APS) (Bio-Rad Laboratories, catalog number: 1610700 )
- Boric acid (ACP Chemicals, catalog number: B2940 )
- EDTA (ACP Chemicals, catalog number: E4320 )
- Tris base (Fisher Scientific, catalog number: BP152 )
- Tris-HCl (Sigma-Aldrich, Roche Diagnostics, catalog number: 10812846001 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- DTT (Sigma-Aldrich, Roche Diagnostics, catalog number: 10197777001 )
- Sodium hydroxide (NaOH) (VWR, BDH, catalog number: BDH9292 )
- Sodium chloride (NaCl) (ACP Chemicals, catalog number: S2830 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Heparin (Sigma-Aldrich, catalog number: H0777-25KU )
- Biotin (Sigma-Aldrich, catalog number: B4501 )
- Yeast extract (Fisher Scientific, catalog number: BP1422 )
- Tryptone (Fisher Scientific, catalog number: BP1421 )
- Potassium hydroxide (KOH) (VWR, BDH, catalog number: BDH9262 )
- 10x Bacterial Native Extract (BNE) buffer (see Recipes)
- 10x Buffer A (see Recipes)
- 10x HEPES KCl (HK) buffer (see Recipes)
- 10x Binding buffer (see Recipes)
- 10x Biotin elution buffer (see Recipes)
- LB media (see Recipes)
Equipment
- Standard personal protective equipment
- Pipettes
- Peristaltic pump
- Fraction collector and appropriate collection tubes
- SpectraMax M5 fluorescent plate reader (Molecular Devices, model: SpectraMax M5 )
- Temperature controlled room (To be maintained at 4 °C)
- Preparative centrifuge, microcentrifuge and benchtop Picofuge
- Tube rotator for 1.5 ml tubes
- Polyacrylamide gel running equipment
- French press
- Sorvall RC6+ centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: SorvallTM RC 6 Plus )
- Sorvall SLA-1500 rotor (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SLA-1500 )
- NanoDrop
- Autoclave
Procedure
- Construct design
- A single copy of RNA Mango is usually sufficient for the pull down analysis and can be inserted into pre-existing stem loops that do not interfere with the function of the RNA of interest (Figure 2 (Panchapakesan et al., 2017; Trachman III et al., 2017). If inserting into a pre-existing stem loop structure, the RNA Mango quadruplex core and GAAA tetraloop-like isolation domain then the Mango aptamer can be inserted within the stem structure using the following sequence: 5’-...NNN NNN GAA GGG ACG GUG CGG AGA GGA GAN’ N’N’N’ N’N’.., where the left arm of the arbitrary RNA helix is indicated by N residues. The right arm (being the reverse complement of the left) is indicated by N’ symbols. The GAAA tetraloop-like isolation motif (shown in italics) together with the TO1-Desthiobiotin binding quadruplex core (in bold) being inserted within the arbitrary helix. While GNRA tetraloops are common in nature, RNA Mango can also be inserted into the 3’ or 5’ end of an unstructured RNA of interest. In this case, RNA Mango (including the GAAA tetraloop-like motif) together with an arbitrary stem should be inserted. New Mango tags (Mango II, III and IV) can be inserted in a similar fashion (Autour et al., 2018).
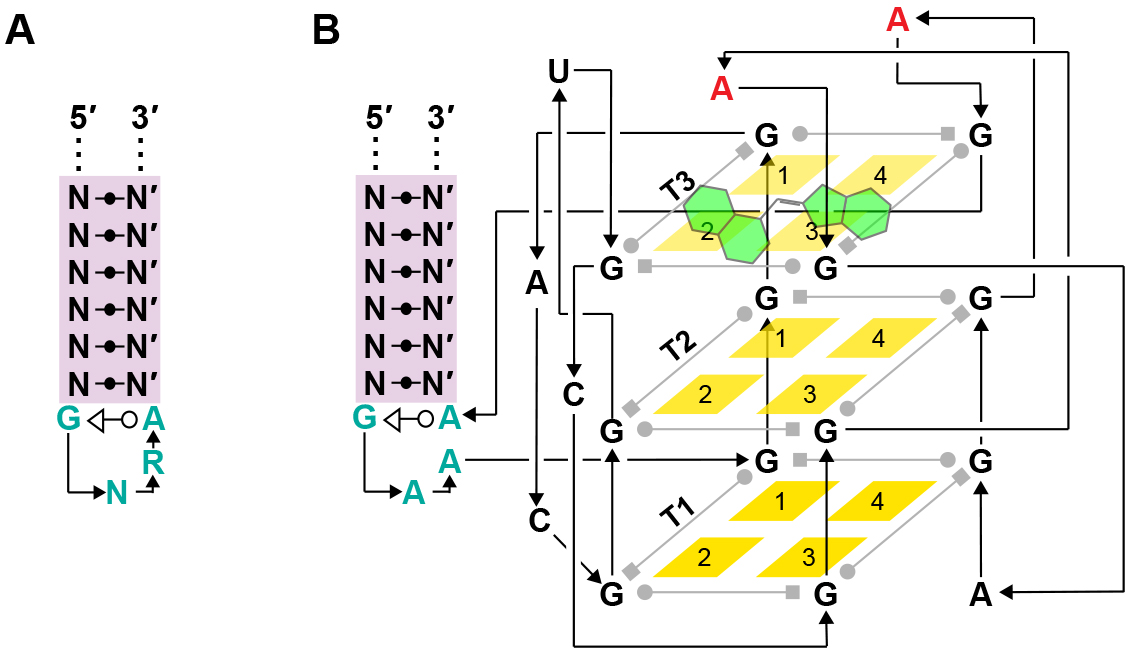
Figure 2. Design of biologically compatible Mango-I tags. A. A general GNRA tetraloop structure is shown, depicted using Leontis-Westhof nomenclature (Leontis and Westhof, 2001). B. Structure of Mango I (figure modified from Autour et al. (2018)). Mango can be incorporated into naturally occurring stem-loop structures by replacing the N-N’ stem highlighted in purple with the stem sequence of the RNA of interest. In Mango I, a GAAA tetraloop-like motif (blue) encloses the core G-quadruplex (3 tiers: T1, T2, T3) by flanking it such that the Mango I core fits into GAA/A, where ‘/’ is the point of the Mango core insertion. Thus, Mango I can be inserted into a pre-existing GNRA tetraloop structure in a similar manner, so long as Mango does not interfere with the tetraloop’s biological function. - The Mango tagged RNA should be tested for the functionalities of the Mango tag as well as the RNA of interest. The functionality of the Mango tag can be tested by measuring the fluorescence of the Mango tagged RNA of interest and comparing it to in vitro transcribed RNA Mango (Panchapakesan et al., 2017). Typically, a 1 µM solution of RNA is made along with two-fold excess of TO1-Dtb ligand in 1x Binding buffer. After incubating at RT for 5 min, fluorescence assays can be performed using a fluorometer. For a SpectraMax M5 fluorometer, the following excitation and emission setup is optimal for analyzing RNA Mango mediated fluorescence: Excitation at 495 nm; Emission 535 nm PMT: Medium, Cut-off 515 nm–Bottom read. Using a SpectraMax M5 under the aforementioned settings, RNA Mango concentrations as low as 5 nM can be reliably detected. The user should optimize the excitation and emission wavelengths to minimize interference from the excitation light source of the specific fluorometer being used.
- A single copy of RNA Mango is usually sufficient for the pull down analysis and can be inserted into pre-existing stem loops that do not interfere with the function of the RNA of interest (Figure 2 (Panchapakesan et al., 2017; Trachman III et al., 2017). If inserting into a pre-existing stem loop structure, the RNA Mango quadruplex core and GAAA tetraloop-like isolation domain then the Mango aptamer can be inserted within the stem structure using the following sequence: 5’-...NNN NNN GAA GGG ACG GUG CGG AGA GGA GAN’ N’N’N’ N’N’.., where the left arm of the arbitrary RNA helix is indicated by N residues. The right arm (being the reverse complement of the left) is indicated by N’ symbols. The GAAA tetraloop-like isolation motif (shown in italics) together with the TO1-Desthiobiotin binding quadruplex core (in bold) being inserted within the arbitrary helix. While GNRA tetraloops are common in nature, RNA Mango can also be inserted into the 3’ or 5’ end of an unstructured RNA of interest. In this case, RNA Mango (including the GAAA tetraloop-like motif) together with an arbitrary stem should be inserted. New Mango tags (Mango II, III and IV) can be inserted in a similar fashion (Autour et al., 2018).
- Native extract preparation
- Bacterial cells (E. coli) with RNA Mango tagged RNA of interest should be grown in appropriate growth conditions to maximize the formation of the RNP complex desired. We typically grew cells overnight in 1 L LB media.
- Cells are then pelleted at 3,800 x g (RCF) for 10 min in a refrigerated Sorvall RC6+ centrifuge, using a Sorvall SLA-1500 rotor. The resulting cell pellet is then re-suspended in 20 ml of pre-chilled BNE buffer in ice. Cells are lysed by passaging through a 4 °C French press twice at 1,000 psi. If a French press is unavailable, then alternative cell disruption techniques such as sonication or enzymatic methods such as lysozyme digestion can be used. This crude lysate is then cleared by centrifugation at 13,200 x g (RCF) for 10 min at 4 °C. The cleared lysate is flash frozen in liquid nitrogen and stored at -80 °C until ready for use. For other organisms, including yeast, appropriate native extract preparation protocols should be used.
- Ensure the presence of KCl (90 to 150 mM), depending upon the stability of RNP complex of interest, as binding to TO1-Desthiobiotin requires KCl (Dolgosheina et al., 2014; Jeng et al., 2016; Panchapakesan et al., 2017). Handle all material in Steps B2 and B3 PROMPTLY on ice.
- Bacterial cells (E. coli) with RNA Mango tagged RNA of interest should be grown in appropriate growth conditions to maximize the formation of the RNP complex desired. We typically grew cells overnight in 1 L LB media.
- TO1-Desthiobiotin streptavidin agarose bead preparation
Prepare an appropriate amount of streptavidin agarose bead slurry. Using 7 ml of bacterial extract and 700 μl of streptavidin agarose bead slurry, derivatized with 42 nanomoles of TO1-Desthiobiotin, we obtained high quality results when overexpressing the 6S regulatory RNA (Dolgosheina et al., 2014; Panchapakesan et al., 2017). Scale the protocol below to suit experimental conditions.- Carefully re-suspend streptavidin agarose bead slurry by gentle agitation.
- Take up 700 μl streptavidin agarose beads in a 1.5 ml Eppendorf tube.
- Wash twice at room temperature (RT) with 1 ml Buffer A, 2 min each, using a standard laboratory rotator. The tubes can be centrifuged briefly in a Picofuge to remove excess liquid from the beads.
- Wash beads three times using 1 ml HK buffer for 2 min each wash, again using a rotator.
- For every 100 μl streptavidin agarose beads incubate with 6 nanomoles of TO1-Desthiobiotin in 100 μl of HK buffer. Incubate at RT for 15 min on a rotator. Note the OD500 (ɛ500 = 63,000 M-1 cm-1) of the fluorophore solution before and after binding to the beads to estimate the amount of fluorophore bound using a NanoDrop. More than 90% of TO1-Desthiobiotin should be bound. The color of the beads will now turn orange because of the fluorophore binding. Different beads may have distinct binding capacities, which should be empirically determined if needed.
- Wash with 1 ml of HK buffer to remove residual unbound fluorophore.
- Carefully re-suspend streptavidin agarose bead slurry by gentle agitation.
- Binding of extract to prepared agarose beads
Since the final goal of this procedure is to purify RNP complexes in native conditions, care should be taken not to denature the complex in every step of the purification viz., bead binding, washing and elution. Since RNA Mango requires KCl for proper folding and binding to the TO1-Desthiobiotin ligand (Dolgosheina et al., 2014; Jeng et al., 2016; Trachman III et al., 2017), KCl should be included in all binding, washing and elution steps. Other components of the buffer can be modified according to experimental needs.- Bind 7 ml of bacterial extract to beads at RT. Incubate at RT for 15 min on a rotator (in the case of yeast extract (Panchapakesan et al., 2017), binding is performed at 4 °C, to ensure stabilization of the RNP complex). The extract can be diluted, if required, by the addition of BNE or Binding buffer to the extract.
- Wash twice with 1 ml Binding buffer for 2 min each on a rotator. The number of washes selected depends on the anticipated stability of the RNP complex. Likewise, the temperature (4-37 °C) and time (2-15 min) of each wash can be significantly adjusted if optimization is required.
- Bind 7 ml of bacterial extract to beads at RT. Incubate at RT for 15 min on a rotator (in the case of yeast extract (Panchapakesan et al., 2017), binding is performed at 4 °C, to ensure stabilization of the RNP complex). The extract can be diluted, if required, by the addition of BNE or Binding buffer to the extract.
- Biotin-mediated native elution
Elute using 20 mM Biotin elution buffer in 500 μl volume at 37 °C (Panchapakesan et al., 2017) for 20 min on a rotator as biotin effectively displaces desthiobiotin from streptavidin (Hirsch et al., 2002). It is desirable to perform downstream processing immediately following biotin-mediated native elution. If not, the sample should be aliquoted and immediately flash frozen in liquid nitrogen for future use. - Downstream processing: Microtiter plate fluorescence assay and gel staining assays
- Load 200 μl of biotin eluate into a 96-well black wall, clear bottom microtiter plate and measure fluorescence emission.
- To estimate the purity, expression level and integrity of the RNA fraction of the Mango pulldown, phenol chloroform extract 20 μl of the biotin eluate, together with equivalent volumes of input, flow through and wash. Perform denaturing 5% PAGE with TBE as described elsewhere. If the RNA is well expressed, SYBR staining can be used to visualize both the RNA of interest and judge levels of RNA contamination. If the RNA expression levels are low, a Northern blot can be used to judge RNA integrity and expression levels.
- Load 200 μl of biotin eluate into a 96-well black wall, clear bottom microtiter plate and measure fluorescence emission.
- Size Exclusion Chromatography (SEC) of streptavidin-purified Mango complex
As the biotin-eluted RNP complex is highly fluorescent (while it is bound to TO1-Desthiobiotin), it can be further purified with secondary purification techniques such as SEC (Panchapakesan et al., 2017). For SEC purification, load onto a size exclusion column containing SEC resin appropriate for the expected molecular size of the RNP complex being purified. To purify the Mango-tagged 6S RNA in complex with bacterial RNAP, we used a Superdex 200 column (24 ml, 16 mm diameter) using HK buffer at 0.1 ml/min flow rate. In this example, 380 μl of the biotin extract was loaded onto the column. The column was run at 4 °C and 500 μl fractions were collected in silanized plastic tubes.- Fraction analysis: 200 μl from each fraction was placed into a 96-well black wall, clear-bottom microtiter plate, and fluorescence measured using a SpectraMax M5 fluorescence plate reader (settings as previously described). Note that RNA Mango fluorescence is temperature sensitive, maximum fluorescence being obtained at lower temperatures (Jeng et al., 2016). Hence, if the signal to noise ratio is lower at RT, the signal can in principle be improved by measuring fluorescence at 4 °C provided that condensation can be avoided.
- Pool the fluorescent fractions of interest. The purified RNP complexes can be analyzed using a variety of techniques such as Mass Spec analysis, Western blot etc. We have used the University of British Columbia Proteomics Core Facility, where samples were analyzed using LC-MS/MS after purification from SDS PAGE gel with success.
- Fraction analysis: 200 μl from each fraction was placed into a 96-well black wall, clear-bottom microtiter plate, and fluorescence measured using a SpectraMax M5 fluorescence plate reader (settings as previously described). Note that RNA Mango fluorescence is temperature sensitive, maximum fluorescence being obtained at lower temperatures (Jeng et al., 2016). Hence, if the signal to noise ratio is lower at RT, the signal can in principle be improved by measuring fluorescence at 4 °C provided that condensation can be avoided.
Data analysis
For more data analysis such as denaturing gel (Step F2), SEC trace of the purified RNP complex (Step G1), the readers may refer to (Dolgosheina et al., 2014; Panchapakesan et al., 2017).
Notes
- KCl is essential for the RNA Mango aptamer to bind TO1-Desthiobiotin, but this binding is only modestly dependent on KCl concentration (Dolgosheina et al., 2014), which can be adjusted as needed (90-150 mM final concentration). However, subsequent generations of Mango (Mango II, III and IV) are much more flexible in optimal KCl concentration range (5-150 mM, Mango III functions optimally at 20 mM KCl) and are functional up to at least 256 mM MgCl2 (Autour et al., 2018).
- We have had success performing purification in the absence of RNase and protease inhibitors, but consider adding these to both the BNE and Binding buffers as needed.
- Although this protocol extensively deals with purification from bacterial cells, it can also be adopted to work with eukaryotic systems such as yeast cells. If using yeast cells, steps such as native extract preparation (Step B1), binding of extract to beads (Procedure D) and elution (Procedure E) should be changed accordingly.
Recipes
Note: Buffers (prepare as 10x sterile filtered stocks, store at -20 °C or lower).
- 10x Bacterial Native Extract (BNE) buffer
200 mM Tris pH 8.0
900 mM KCl
10 mM MgCl2
Prepare a 100 mM DTT stock and make up 1x BNE buffer to include 1 mM fresh DTT (final) before use - 10x Buffer A
1,000 mM NaOH
500 mM NaCl - 10x HEPES KCl (HK) buffer
150 mM HEPES pH 7.5
900 mM KCl - 10x Binding buffer
150 mM HEPES pH 7.5
900 mM KCl
750 μg/ml heparin*
Prepare 1x Binding buffer from HK buffer by adding 1 mM fresh DTT (final) immediately before use. As many RNA binding proteins are positively charged, heparin (a negatively charged sulfated carbohydrate) may be added as a cheap competitor to inhibit nonspecific RNP complex formation - 10x Biotin elution buffer
200 mM Biotin in 10x Binding buffer - LB media
10 g tryptone
5 g yeast extract
10 g NaCl
Autoclave prior to use
Acknowledgments
PJU acknowledges an NSERC Discovery operating grant. The authors declare no conflicts of interest or competing interests.
References
- Autour, A., S, C. Y. J., A, D. C., Abdolahzadeh, A., Galli, A., Panchapakesan, S. S. S., Rueda, D., Ryckelynck, M. and Unrau, P. J. (2018). Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat Commun 9(1): 656.
- Dolgosheina, E. V., Jeng, S. C., Panchapakesan, S. S., Cojocaru, R., Chen, P. S., Wilson, P. D., Hawkins, N., Wiggins, P. A. and Unrau, P. J. (2014). RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol 9(10): 2412-2420.
- Hirsch, J. D., Eslamizar, L., Filanoski, B. J., Malekzadeh, N., Haugland, R. P., Beechem, J. M. and Haugland, R. P. (2002). Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal Biochem 308(2): 343-357.
- Jeng, S. C., Chan, H. H., Booy, E. P., McKenna, S. A. and Unrau, P. J. (2016). Fluorophore ligand binding and complex stabilization of the RNA Mango and RNA Spinach aptamers. RNA 22(12): 1884-1892.
- Leontis, N. B. and Westhof, E. (2001). Geometric nomenclature and classification of RNA base pairs. RNA 7(4): 499-512.
- Panchapakesan, S. S. S., Ferguson, M. L., Hayden, E. J., Chen, X., Hoskins, A. A. and Unrau, P. J. (2017). Ribonucleoprotein purification and characterization using RNA mango. RNA 23(10): 1592-1599.
- Panchapakesan, S. S., Jeng, S. C. and Unrau, P. J. (2015). RNA complex purification using high-affinity fluorescent RNA aptamer tags. Ann N Y Acad Sci 1341: 149-155.
- Trachman, R. J., 3rd, Demeshkina, N. A., Lau, M. W. L., Panchapakesan, S. S. S., Jeng, S. C. Y., Unrau, P. J. and Ferre-D’Amare, A. R. (2017). Structural basis for high-affinity fluorophore binding and activation by RNA mango. Nat Chem Biol 13(7): 807-813.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Panchapakesan, S. S. S., Jeng, S. C. Y. and Unrau, P. J. (2018). Purification of RNA Mango Tagged Native RNA-protein Complexes from Cellular Extracts Using TO1-Desthiobiotin Fluorophore Ligand. Bio-protocol 8(7): e2799. DOI: 10.21769/BioProtoc.2799.
Category
Microbiology > Microbial biochemistry > RNA
Biochemistry > RNA > RNA-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









