- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA Purification from the Unicellular Green Alga, Chromochloris zofingiensis
Published: Vol 8, Iss 7, Apr 5, 2018 DOI: 10.21769/BioProtoc.2793 Views: 8150
Reviewed by: Adam IdoineClaudia CatalanottiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Nuclei in Tagged Cell Types (INTACT), RNA Extraction and Ribosomal RNA Degradation to Prepare Material for RNA-Seq
Mauricio A. Reynoso [...] Kaisa Kajala
Apr 5, 2018 16205 Views

Extraction of RNA from Recalcitrant Tree Species Paulownia elongata
Niveditha Ramadoss and Chhandak Basu
Jul 20, 2018 7716 Views

Simple Method for Efficient RNA Extraction From Arabidopsis Embryos
Fernanda Marchetti [...] Eduardo Zabaleta
Feb 20, 2025 1914 Views
Abstract
Chromochloris zofingiensis is a unicellular green alga that is an emerging model species for studies in fields such as biofuel production, ketocarotenoid biosynthesis and metabolism. The recent availability of a high-quality genome assembly facilitates systems-level analysis, such as RNA-Seq. However, cells of this alga have a tough cell wall, which is a challenge for RNA purification. This protocol was designed specifically to breach the cell wall and isolate high-quality RNA suitable for RNA-Seq studies.
Keywords: Chromochloris zofingiensisBackground
Chromochloris zofingiensis is a small unicellular green alga from the chlorophyte lineage (Dönz, 1934). Previously, this species has been described in the literature with the genera Muriella, Chlorella, and Mychonastes (Fučíková and Lewis, 2012). There are strong economic interests in C. zofingiensis because it is capable of producing large quantities of lipids for biofuels and shows promise as a source of the commercially valuable nutraceutical astaxanthin (Breuer et al., 2012; Mulders et al., 2014; Liu et al., 2016). Recently, a high-quality, chromosome-level genome assembly and accompanying annotations were published, which facilitates systems level analyses like RNA-Seq (Roth et al., 2017). The following protocol was designed to produce highly purified total RNA, including miRNA, from liquid cultures of C. zofingiensis suitable for RNA-Seq. C. zofingiensis cells are protected by a robust cell wall, which this protocol was designed to penetrate. Starting material for the protocol can be up to 2.5 x 108 total cells. This protocol should yield at least 20 µg of highly purified RNA suitable for the preparation of RNA-Seq libraries by standard kits, such as the Illumina TruSeq Stranded Total RNA kit.
Materials and Reagents
- 50 ml Falcon tubes (Corning, Falcon®, catalog number: 352070 )
- Serological pipettes, 10 ml (Fisher Scientific, catalog number: 13-676-10J )
- Lysing Matrix D Tubes (2 ml) (MP Biomedicals, catalog number: 116913050 )
- Tapered end, metal spatula (Fisher Scientific, catalog number: 14-374 )
- VacConnectors (QIAGEN, catalog number: 19407 )
- C. zofingiensis (SAG, catalog number: 211-14 )
- Proteose medium (UTEX Culture Collection of Algae, Proteose medium)
- Chu’s medium (UTEX Culture Collection of Algae, Chu’s medium)
- Liquid nitrogen (LN2) (various)
- Wet ice (various)
- Dry ice (various)
- RNaseZap (250 ml spray bottle) (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9780 )
- RNase-free ethanol (EtOH) (100% and 70%) (various)
- TRIzol (100 ml) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15596026 )
- Chloroform:isoamyl alcohol (24:1) (Sigma-Aldrich, catalog number: C0549-1PT )
- MaXtract High Density (100 x 15 ml) (QIAGEN, catalog number: 129065 )
- miRNeasy Mini Kit (50) (QIAGEN, catalog number: 217004 ), which includes:
- QIAGEN miRNeasy mini columnsa.QIAGEN miRNeasy mini columns
- 1.5 and 2 ml collection tubes
- QIAGEN buffer RWT
- QIAGEN buffer RPE
- RNase-free water
- QIAGEN miRNeasy mini columnsa.QIAGEN miRNeasy mini columns
- RNase-Free DNase Set (50 samples) (QIAGEN, catalog number: 79254 ), which includes:
- RNase-free DNase I
- QIAGEN buffer RDD
- RNase-free DNase I
- 3 M sodium acetate (NaOAc), pH = 8, RNase-free (various)
- 1 M Tris hydrochloride (Tris-HCl), pH = 8, RNase-free (various)
- 5 M sodium chloride (NaCl), RNase-free (various)
- 0.5 M ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA), pH = 8, RNase-free (various)
- 10% sodium dodecyl sulfate (SDS) (various)
- 20 mg/ml Proteinase K (Fisher Scientific, catalog number: BP1700-100 )
- Agilent RNA 6000 Nano kit (Agilent Technologies, catalog number: 5067-1511 )
- Lysis buffer (see Recipes)
Equipment
- Refrigerated centrifuge (Eppendorf, model: 5810 R )
- Refrigerated microfuge (Eppendorf, model: 5424 R )
- Clean bench
- Ice buckets (Corning, catalog number: 432122 )
- PIPETMAN Classic pipet (Gilson, model: P20, catalog number: F123600 )
- PIPETMAN Classic pipet (Gilson, model: P200, catalog number: F123601 )
- PIPETMAN Classic pipet (Gilson, model: P1000, catalog number: F123602 )
- Homogenizer (MP Biomedicals, model: FastPrep-24TM 5G )
- Homogenizer adaptor (MP Biomedicals, model: CoolPrepTM 24x2mL )
- Vacuum manifold (QIAGEN, model: QIAvac 24 Plus )
- Heat block (VWR, catalog number: 75838-286 )
- Spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 )
- Bioanalyzer (Agilent Technologies, model: 2100 Bioanalyzer )
Procedure
- Cell collection
This protocol is designed for use on ~2.5 x 108 cells or less (e.g., 50 ml of culture at 5 x 106 cells/ml density) of liquid cultures of C. zofingiensis. See Figure 1 for example.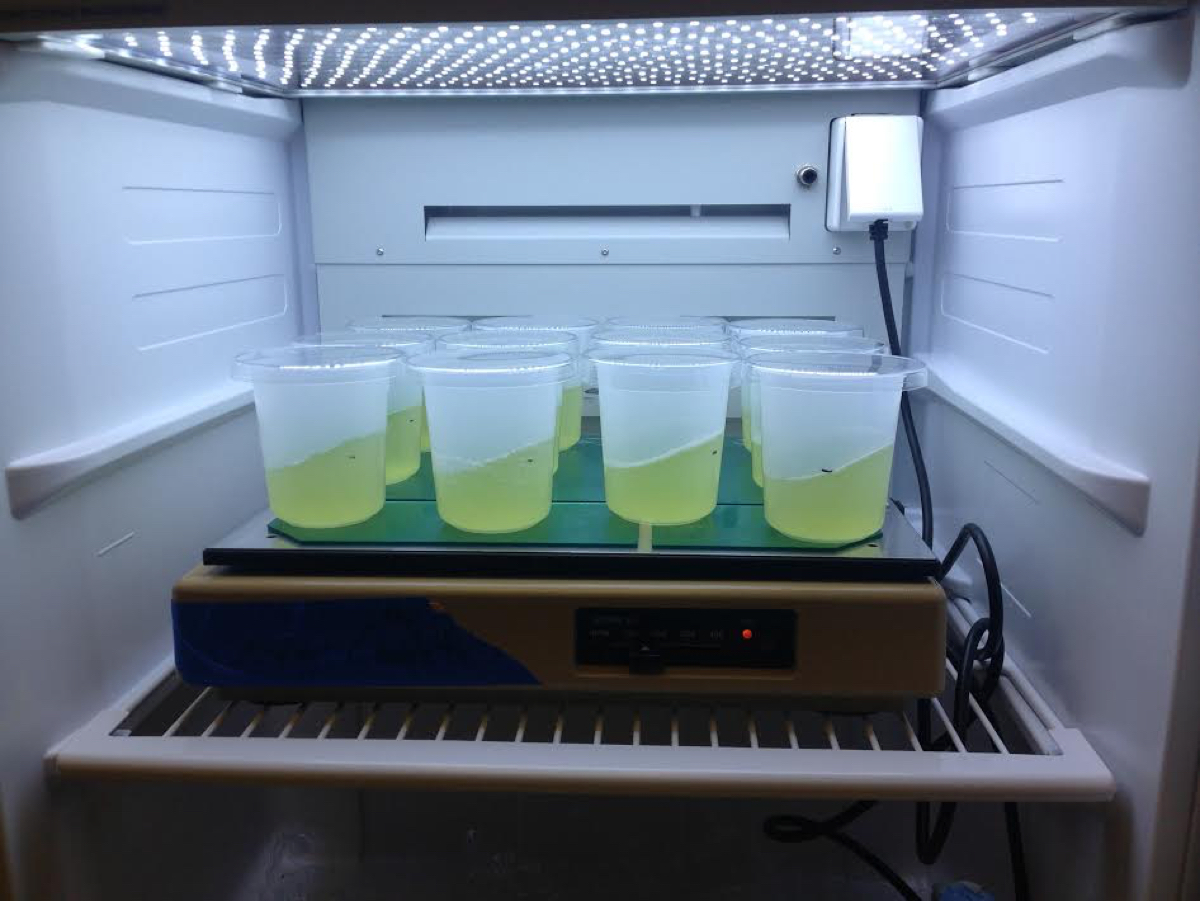
Figure 1. Liquid cultures of C. zofingiensis. An example of liquid cultures of C. zofingiensis (SAG 211-14) suitable as input for this protocol. Here, cells were grown in Proteose medium (UTEX Culture Collection of Algae) supplemented with Chu’s medium (2 ml/L, UTEX Culture Collection of Algae) as described previously (Roth et al., 2017). The cultures were grown at 25 °C with shaking at 100-150 rpm in diurnal (16 h light, 8 h dark) conditions with a light intensity of 90-100 μmol photons m-2 sec-1.- Transfer cell culture to a 50 ml Falcon tube.
- Spin for 5 min at 3,200 x g (RCF) at 4 °C. Immediately after the centrifuge comes to a stop, quickly remove the supernatant by decanting into a waste receptacle.
- Flash freeze tubes containing cell pellet in liquid N2.
- Transfer cell culture to a 50 ml Falcon tube.
- Preparation
- Prepare glassware and metalware including spatula by washing with detergent and baking at 180 °C for 8 h.
- Clean bench and pipettes with RNaseZap or equivalent.
- Prepare fresh lysis buffer (see Recipes) just prior to starting according to the recipe.
- Pre-cool microfuge, centrifuge and buckets to 4 °C.
- Prepare buckets with crushed wet ice.
- Prepare buckets with crushed dry ice.
- Chill 100% EtOH on dry ice.
- Label Lysing Matrix D tubes and place on dry ice.
- Prepare glassware and metalware including spatula by washing with detergent and baking at 180 °C for 8 h.
- Cell lysis and RNA purification
- Transfer frozen sample tubes to the dry ice bucket.
- Take a clean, RNase-free spatula and thrust it up and down into the tube to break up the cell pellet. The tube should remain on dry ice during the process so that the cells remain frozen and prevent RNA degradation.
- Pipette 1 ml of cold 100% EtOH into the tube.
- Transfer the cell mass with spatula and EtOH with a pipette into a chilled Lysing Matrix D 2 ml tube.
- Spin for 3 min at 2,700 x g (RCF) at 4 °C. Decant supernatant.
- Place crushed dry ice in the homogenizer tray, transfer tubes to the homogenizer, and run for 60 sec at 6.0 m/sec. Repeat for an additional 60 sec.
- Add 1 ml of freshly prepared lysis buffer to the cell mass.
- Vortex for ~30 sec.
- Incubate for 3 min at RT.
- Spin for 3 min at 20,800 x g (RCF) at RT.
- Spin MaXtract tubes for 2 min at 1,500 x g (RCF) at RT.
- Add 10 ml of TRIzol to the MaXtract tube.
- Transfer 1 ml of supernatant to the MaXtract tube with TRIzol.
- Incubate for 3 min at RT.
- Add 2 ml of chloroform:isoamyl alcohol, and shake vigorously for 30 sec.
- Incubate for 5 min at RT.
- Centrifuge for 5 min at 1,500 x g (RCF) at RT.
- Add 10 ml of cold 100% EtOH to a labeled 50 ml conical tube for each sample.
- Decant the aqueous phase of the MaXtract tube (~6-7 ml) into the 50 ml conical tube with EtOH.
- Invert the tube several times to mix, and then place on wet ice.
- Working with one sample at a time, while the others remain on ice:
- Turn on the vacuum. Place a QIAGEN miRNeasy mini-column on the vacuum manifold with a fresh VacConnector.
- Using a 10 ml serological pipette, load the lysate/EtOH mixture onto the miRNeasy mini column until all liquid has passed through the membrane.
- Close vacuum and place column back in its 2 ml collection tube and put on ice.
- Turn on the vacuum. Place a QIAGEN miRNeasy mini-column on the vacuum manifold with a fresh VacConnector.
- Put column back on vacuum manifold and add 350 µl QIAGEN buffer RWT. Open vacuum until all buffer has passed through the column.
- Return the column to the collection tube.
- Just prior to using, prepare fresh DNase solution from the QIAGEN RNase-free DNase Set. Combine sufficient DNase (10 µl per column) and QIAGEN buffer RDD (70 µl per column) for all samples. Mix gently by inverting the tube 3-4 times.
Note: Do NOT vortex the DNase! - Add 80 µl DNase/RDD mixture from the previous step to each column.
- Incubate tubes for 15 min in a 30 °C heat block.
- Place columns back on the vacuum manifold.
- Add 350 µl QIAGEN buffer RWT to each column.
- Open vacuum until all RWT buffer has passed through the columns.
- Wash columns with QIAGEN buffer RPE buffer twice as follows:
- Add 500 µl of RPE to each column.
- Close the lid and rotate the column gently to rinse the walls of the column.
- Place columns back on the vacuum manifold.
- Open vacuum until all RPE buffer has passed through the columns.
- Repeat.
- Add 500 µl of RPE to each column.
- Carefully remove the column from the collection tube to avoid carryover of ethanol and place it in a new 2 ml collection tube.
- Spin for 1 min at maximum speed (≥ 10,000 x g (RCF)) at RT.
- Place the column in a new, labeled 1.5 ml collection tube.
- Add 50 µl RNase-free dH2O directly to the spin column membrane.
- Spin for 1 min at maximum speed (≥ 10,000 x g (RCF)) at RT to elute the RNA.
- Repeat with an additional 50 µl RNase-free dH2O (~90 µl eluate in total).
- Add 270 µl RNase-free dH2O.
- Add 40 µl 3 M NaOAc at pH 8. Recap and invert the tube to mix.
- Add 1,000 µl ice-cold 100% EtOH. Recap and invert the tube to mix.
- Incubate for 30 min at -20 °C.
- Spin for 30 min at 4 °C at maximum speed (≥ 10,000 x g (RCF)).
- Without disturbing the pellet, gently decant supernatant.
- Wash with 500 µl RT 70% EtOH.
- Without disturbing the pellet, gently decant supernatant. Remove remaining liquid with a Pipetman.
- Air dry the pellet for 5-10 min at RT.
- Resuspend the pellet in 60 µl RNase-free dH2O.
- Transfer frozen sample tubes to the dry ice bucket.
- Quality control
- Determine the optical absorbance by NanoDrop spectrophotometer. Use 1 µl of dH2O to blank the NanoDrop.
- Determine the optical absorbance at 260, 230, and 280 nm for 1 µl of each sample. Assay the RNA quality by microfluidic electrophoresis on a Bioanalyzer.
- Dilute 1 µl of each sample with 9 µl of RNase-free water.
- Following the manufacturer’s protocol, load 1 µl of the 10x diluted RNA into a well of an RNA 6000 Nano chip and assay with the Eukaryote total RNA program.
Note: Alternatively, an Agilent Tape Station may be used.
- Dilute 1 µl of each sample with 9 µl of RNase-free water.
- Determine the optical absorbance by NanoDrop spectrophotometer. Use 1 µl of dH2O to blank the NanoDrop.
Data analysis
- Review the NanoDrop spectrophotometer data
- The A260/A280 ratio should be approximately 2.1. Discard sample if ratio is < 2.0 or > 2.2.
- The A260/A230 ratio should be approximately 2.1-2.4. Discard sample if ratio is < 2.0.
- The A260/A280 ratio should be approximately 2.1. Discard sample if ratio is < 2.0 or > 2.2.
- Review the Bioanalyzer report
- There should be prominent peaks at ~1,800 nt and ~3,200 nt for the 18S and 28S, respectively (Figure 2).
- The RIN value calculated by the Agilent software should be > 8. It is recommended that samples with RIN < 7 should be discarded.
Note: RIN values calculated by the software may be lower than similar quality RNA from other species due to abundant plastid rRNA in C. zofingiensis preparations.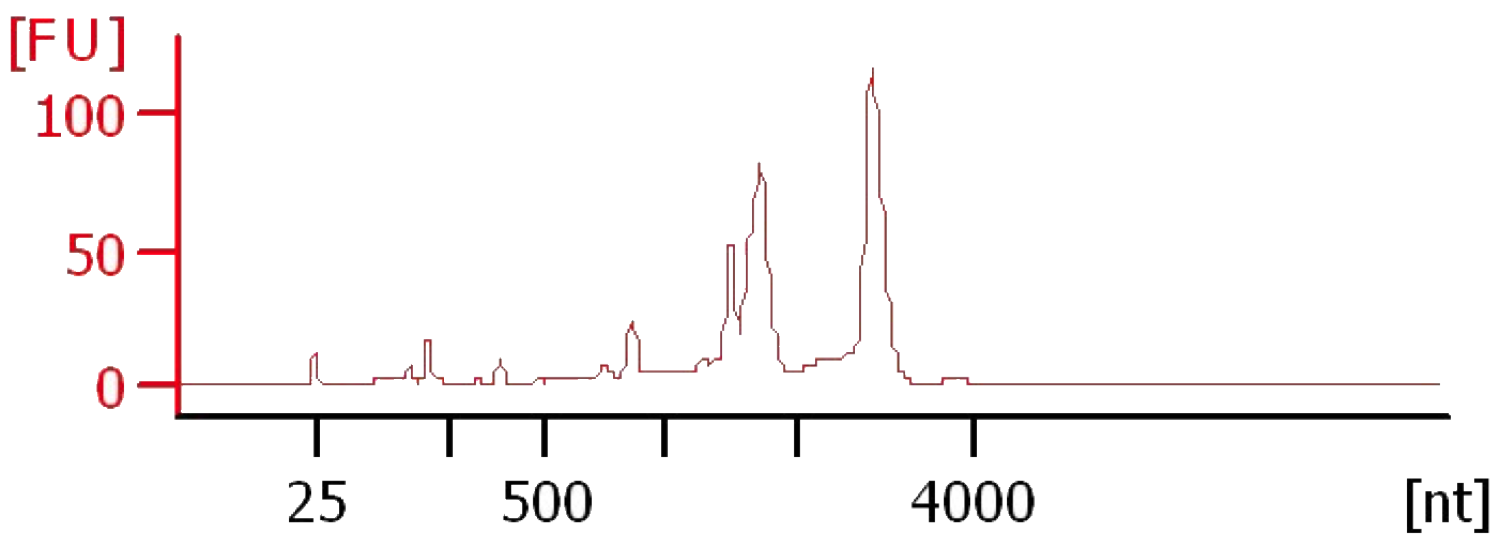
Figure 2. Electropherogram of C. zofingiensis RNA. Total RNA from C. zofingiensis was prepared by the accompanying protocol and ran on an Agilent 2100 BioAnalyzer with a Eukaryote Total RNA Pico chip.
- There should be prominent peaks at ~1,800 nt and ~3,200 nt for the 18S and 28S, respectively (Figure 2).
Notes
RNA is extremely sensitive to damage by RNase. Careful laboratory hygiene is crucial for minimizing this risk. Additionally, cells must be kept frozen until lysis buffer is added to prevent RNA degradation. When preparing many RNA samples, some may not satisfy the quality control criteria specified in the Data analysis section. It may be prudent to prepare more replicate samples than your experiment requires so that some may be discarded at the end of this protocol without compromising the overall study.
Recipes
- Lysis buffer
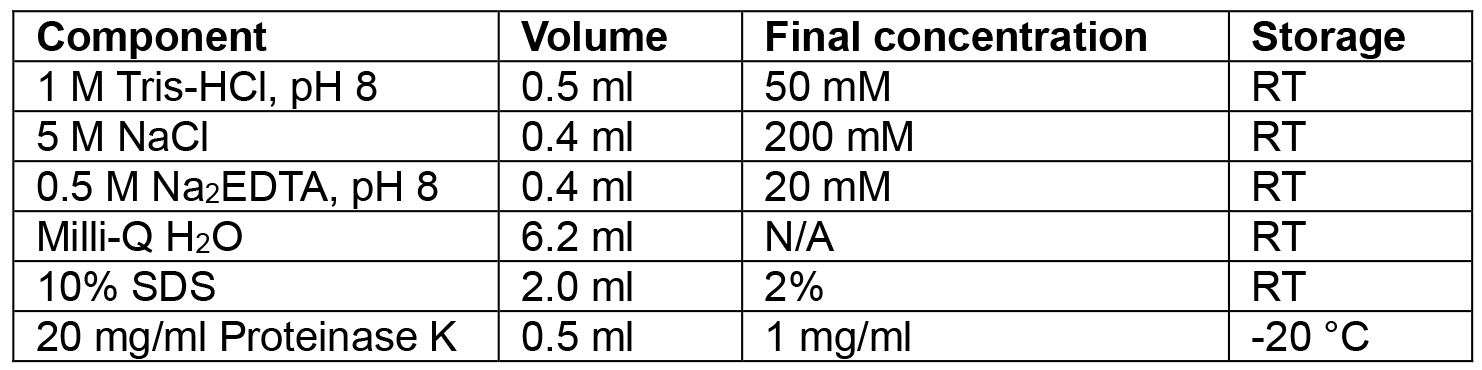
Volumes given are for 10 ml. Adjust up or down as needed
Add the SDS and Proteinase K in just before use
Acknowledgments
This protocol has been adapted from a previously published paper (Roth et al., 2017). This work was supported by the Agriculture and Food Research Initiative Competitive Grant No. 2013-67012-21272 from the USDA National Institute of Food and Agriculture (to M.S.R), and by a cooperative agreement with the US Department of Energy Office of Science, Office of Biological and Environmental Research program under Award DE-FC02-02ER63421. The authors declare no conflicts of interest or competing interests.
References
- Breuer, G., Lamers, P. P., Martens, D. E., Draaisma, R. B. and Wijffels, R. H. (2012). The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour Technol 124: 217-226.
- Dönz, O.C. (1934). Chlorella zofingiensis, eine neue Bodenalge. Ber Schweiz Bot Ges 43: 127-131.
- Fučíková, K. and Lewis, L. A. (2012). Intersection of Chlorella, Muriella and Bracteacoccus: Resurrecting the genus Chromochloris Kol et Chodat (Chlorophyceae, Chlorophyta). Fottea 12: 83-93.
- Liu, J., Mao, X., Zhou, W. and Guarnieri, M. T. (2016). Simultaneous production of triacylglycerol and high-value carotenoids by the astaxanthin-producing oleaginous green microalga Chlorella zofingiensis. Bioresour Technol 214: 319-327.
- Mulders, K. J. M., Janssen, J. H., Martens, D. E., Wijffels, R. H. and Lamers, P. P. (2014). Effect of biomass concentration on secondary carotenoids and triacylglycerol (TAG) accumulation in nitrogen-depleted Chlorella zofingiensis. Algal Res 6: 8-16.
- Roth, M. S., Cokus, S. J., Gallaher, S. D., Walter, A., Lopez, D., Erickson, E., Endelman, B., Westcott, D., Larabell, C. A., Merchant, S. S., Pellegrini, M. and Niyogi, K. K. (2017). Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proc Natl Acad Sci U S A 114(21): E4296-E4305.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gallaher, S. D. and Roth, M. S. (2018). RNA Purification from the Unicellular Green Alga, Chromochloris zofingiensis. Bio-protocol 8(7): e2793. DOI: 10.21769/BioProtoc.2793.
Category
Plant Science > Plant molecular biology > RNA > RNA extraction
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










