- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bacterial Competition Assay Based on Extracellular D-amino Acid Production
Published: Vol 8, Iss 7, Apr 5, 2018 DOI: 10.21769/BioProtoc.2787 Views: 9191
Reviewed by: Emily CopeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection of D-glutamate Production from the Dual Function Enzyme, 4-amino-4-deoxychorismate Lyase/D-amino Acid Transaminase, in Mycobacterium smegmatis
Helen K. Opel-Reading [...] Kurt L. Krause
Jan 5, 2019 5802 Views

Growth Recovery Assay and FACS-based Population Sorting Following Territorial Exclusion in Proteus mirabilis
Murray J. Tipping and Karine A. Gibbs
Mar 5, 2020 5072 Views

A High-throughput Interbacterial Competition Platform
Hsiao-Han Lin and Erh-Min Lai
Sep 5, 2020 5914 Views
Abstract
Bacteria live in polymicrobial communities under tough competition. To persist in a specific niche many species produce toxic extracellular effectors as a strategy to interfere with the growth of nearby microbes. One of such effectors are the non-canonical D-amino acids. Here we describe a method to test the effect of D-amino acid production in fitness/survival of bacterial subpopulations within a community. Co-cultivation methods usually involve the growth of the competing bacteria in the same container. Therefore, within such mixed cultures the effect on growth caused by extracellular metabolites cannot be distinguished from direct physical interactions between species (e.g., T6SS effectors). However, this problem can be easily solved by using a filtration unit that allows free diffusion of small metabolites, like L- and D-amino acids, while keeping the different subpopulations in independent compartments.
With this method, we have demonstrated that D-arginine is a bactericide effector produced by Vibrio cholerae, which strongly influences survival of diverse microbial subpopulations. Moreover, D-arginine can be used as a cooperative instrument in mixed Vibrio communities to protect non-producing members from competing bacteria.
Background
Bacteria live in polymicrobial communities where a great diversity of species coexist and compete for the available resources. One of the many tactics that bacteria have devised to persist in a specific niche is the production of toxic extracellular metabolites as a strategy to interfere with growth and/or viability of other microbes. D-amino acids have been known for a long time to have a powerful effect in cell shape and viability in bacterial cultures (Bopp, 1965; Fox et al., 1944; Kobayashi et al., 1948; Yaw and Kakavas, 1952; Lark and Lark, 1959; Grula, 1960; Tuttle and Gest, 1960). However, it has not been until recently that D-amino acids have gained physiological meaning when it was reported that many taxonomically unrelated bacteria could release millimolar concentrations of non-canonical D-amino acids (NCDAAs) to the extracellular medium (Lam et al., 2009). Vibrio cholerae, the causative agent of the diarrheal disease cholerae, presents a periplasmic broad spectrum racemase called BsrV reported to produce a great variety of D-amino acids, mainly D-Met and D-Leu (Lam et al., 2009; Cava et al., 2011). Further studies demonstrated that the main mode of action of these D-amino acids was through their incorporation into the peptidoglycan polymer, an essential bacterial structure that plays a role in morphology determination and cell integrity (Caparros et al., 1992; Lam et al., 2009; Cava et al., 2011). Peptidoglycan is a macromolecule composed of glycan chains crosslinked by short peptides. Interestingly, NCDAAs can be incorporated into the peptidoglycan into the 4th or the 5th residue of the peptide stem of the muropeptide subunits and this editing has a key role in synchronizing cell wall metabolism with growth arrest (Lam et al., 2009; Cava et al., 2011).
A recent study demonstrated that the cell wall is not the only target of non-canonical D-amino acids (Alvarez et al., 2018). V. cholerae and many other bacteria produce a great variety of D-amino acids which have distinct functions (Lam et al., 2009; Alvarez et al., 2018). D-arginine stands out as a fitness modulator of bacterial subpopulations, since it shows a significantly higher growth inhibitory activity against a wide diversity of bacterial species compared with other D-amino acids. In contrast to D-methionine, which has a major modulatory role in cell wall biosynthesis, D-arginine growth inhibition is suppressed by mutations in the chaperone systems and the phosphate uptake machinery in several model organisms, strongly supporting different roles for NCDAAs in bacterial physiology (Alvarez et al., 2018).
Co-cultivation is an excellent method to assess the inhibitory effect of D-arginine in mixed bacterial populations. However, when the competing bacteria present very different growth rates (e.g., V. cholerae and Caulobacter crescentus used in this study), relative cell counting can be challenging. Besides, it might be difficult to assess the role of small metabolites in species competition when other mechanisms, such as cell-to-cell dependent interactions (e.g., T6SS), can occur simultaneously. Here we present a method to assess the effect of small metabolites on bacterial populations. The design is based in the compartmentalization of the competing subpopulations in two independent rooms separated by a filter that permits diffusion of small metabolites such as amino acids. Furthermore, this method can be used to demonstrate the metabolic cooperation between producer and non-producer bacteria (e.g., V. cholerae wild-type and ΔbsrV mutant) that share extracellular D-amino acids to outcompete other species in the environment. Finally, we also describe the methodology to determine the total D-amino acid concentration in the media.
Materials and Reagents
- Wired-loop or disposable inoculation loops (SARSTEDT, catalog number: 86.1562.050 )
- 15 ml test tubes (SARSTEDT, catalog number: 62.554.502 )
- Cuvettes (SARSTEDT, catalog number: 67.742 )
- 150 ml Stericup filtration units, 0.22 µm pore size (Merck, catalog number: SCGPU01RE )
- Adhesive tape
- Parafilm (Sigma-Aldrich, catalog number: P7793-1EA )
- Needles (BD, catalog number: 302200 )
- Syringes 1 ml, 10 ml (BD, catalog numbers: 303172 , 307736 )
- 1.5 ml microtubes (Eppendorf, catalog number: 0030120086 )
- Sterile clear flat-bottom 96-well plates with lid (Corning, Falcon®, catalog number: 353072 )
- Sterile glass beads 3 mm (Merck, catalog number: 1040150500 )
- Petri dishes (SARSTEDT, catalog number: 82.1473 )
- Filter units, 0.22 µm pore size (Merck, catalog number: SLGS033SB )
- Disposable pipette tips (VWR, catalog numbers: 613-1083 , 613-1079 , 613-1077 )
- Bacterial strains: V. cholerae N16961 lacZ+ wild-type, V. cholerae N16961 lacZ- ΔbsrV, Caulobacter crescentus NA1000
- Trigonopsis variabilis DAAO (gift from Jose M. Guisan, Catalysis Department, ICP – CSIC, Spain) (Komarova et al., 2012)
- L-Arginine (L-Arg) (Sigma-Aldrich, catalog number: A5006-100G )
- D-Arginine (D-Arg) (Sigma-Aldrich, catalog number: A2646-5G )
- Distilled water
- MilliQ water
- Hydrochloric acid fuming 37% (HCl) (Merck, catalog number: 1003171000 )
- Tryptone (Peptone from casein) (VWR, catalog number: 84610.0500 )
- Yeast extract (VWR, catalog number: 84601.0500 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 71376-1KG )
- Sodium hydroxide pellets (NaOH) (Merck, catalog number: 1064821000 )
- Peptone, meat (enzymatic digest of animal tissue) (VWR, catalog number: 84620.0500 )
- Magnesium sulfate (MgSO4) (Sigma-Aldrich, catalog number: M2643-500G )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C5670-500G )
- Bacteriological agar (VWR, catalog number: 84609.0500 )
- Sodium phosphate monobasic monohydrate (NaH2PO4·H2O) (Sigma-Aldrich, catalog number: 71507-250G )
- Sodium phosphate dibasic heptahydrate (Na2HPO4·7H2O) (Sigma-Aldrich, catalog number: 431478-250G )
- Ortho-phosphoric acid 85% (Merck, catalog number: 1005731000 )
- Flavin adenine dinucleotide disodium salt hydrate (FAD) (Sigma-Aldrich, catalog number: F6625-100MG )
- o-Phenylenediamine (OPD) (Sigma-Aldrich, catalog number: P23938-5G )
- Methanol (VWR, catalog number: 20847.307 )
- Horseradish peroxidase (Sigma-Aldrich, catalog number: 77332-100MG )
- LB medium (see Recipes)
- PYE medium (see Recipes)
- Agar plates (see Recipes)
- L- and D-amino acid stock solutions (see Recipes)
- Sodium phosphate buffer 500 mM pH 7.5 (see Recipes)
- DAAO reaction buffer (see Recipes)
Equipment
- Laminar flow cabinet
- Bunsen burner
- Pipettes (Gilson, catalog numbers: F144563 , F144565 , F144566 )
- Multichannel pipettes (Gilson, catalog number: F14403 )
- Glassware: bottles, measurement cylinders, beakers
- pH-meter (VWR, catalog number: 662-1422 )
- Autoclave (CertoClav, catalog number: 8510174 )
- Incubator (Memmert, catalog number: IN55 )
- Shaker incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: MaxQTM 5000, catalog number: SHKE5000 )
- Thermomixer with adapter for multi-well plates (Eppendorf, catalog numbers: 5355000011 , 5363000012 )
- Spectrophotometer (GE Healthcare, catalog number: 29003605 )
- Microplate reader (Biotek, model: EONTM, catalog number: EONC )
Procedure
- Bacterial co-cultivation
Note: All steps need to be performed under sterile conditions, working by the flame or inside a laminar flow cabinet.- Streak V. cholerae cells from the freezer stock onto an LB agar plate and incubate upside-down for 16 h at 37 °C. V. cholerae lacZ+ wild-type and V. cholerae lacZ- ΔbsrV can be used to assess the role of BsrV in D-amino acid production.
- Streak C. crescentus cells from the freezer stock onto a PYE agar plate and incubate upside-down for 48 h at 28 °C.
Note: Any combination of bacteria (ideally one of them a D-amino acid producer) can be used in this protocol, as long as both can grow in compatible conditions. - Pick a single colony of each strain from the agar plates with a sterile wired-loop and inoculate 2 ml liquid PYE medium in a 15 ml test tube. Grow liquid bacterial cultures overnight (16-18 h) at 28 °C, with shaking at 150 rpm.
Note: Temperature and shaking may vary depending on the bacterial species used in the competition assay. - Determine the OD600 of the cultures: dilute 100 µl overnight culture in 900 µl fresh PYE medium, transfer to a cuvette and read absorbance at 600 nm using a spectrophotometer. Use a cuvette filled with 1 ml fresh PYE as blank.
- Dilute the bacterial suspension in 10 ml PYE medium to a starting concentration of OD600 = 0.01. Depending on the bacterial strain used, the OD600 to CFU ml-1 ratio will differ and needs to be determined for each strain: for example, for the V. cholerae wild type strain, OD600 0.01 = ~107 cells ml-1.
- Stericup filtration units are used as co-cultivation chambers. Fill the upper part, which we called compartment A, with 100 ml fresh PYE medium. Fill the lower part, which we called compartment B, with 200 ml fresh PYE medium. Tightly attach both compartments. Use Parafilm and adhesive tape to firmly seal the lid of compartment A and avoid leakage and place the co-cultivation chamber horizontally. With a hot needle, make a small hole in the top of each compartment to allow inoculation and sample collection; this hole can be sealed with adhesive tape (Figure 1A).
Note: Make sure the co-cultivation chambers are placed horizontally with the hole facing the top, to minimize sample spilling. Here we use 100 ml medium in compartment A and 200 ml medium in compartment B because the size of both parts is different; ideally both compartments should have the same size, shape and medium volume. - L-amino acid supplementation: with a 10 ml syringe and a needle, add 2.5 ml and 5 ml L-arginine (200 mM sterile stock solution) to compartments A and B, respectively (final L-amino acid concentration is 5 mM in each compartment). Use non-supplemented co-cultivation chambers to assess growth in absence of L- or D-amino acids (Figure 1B).
- With a 1 ml syringe and a needle, inoculate the bacteria into the co-cultivation chambers: 100 µl V. cholerae (OD600 = 0.01) is inoculated into compartment A; 500 µl C. crescentus (OD600 = 0.01) is inoculated into compartment B. Use non-inoculated chambers as a control of contamination and growth in absence of a competitor bacteria (Figure 1B).
Note: Determination of the growth rate of the bacteria in competition should be assessed beforehand, including the growing condition (medium and temperature) compatibility. We use 2.5-fold C. crescentus inoculum to compensate for the very different growth rates of V. cholerae and C. crescentus in PYE medium. V. cholerae lacZ+ wild-type is used as D-amino acid producing bacteria. V. cholerae lacZ- ΔbsrV is used as D-amino acid non-producer. A 1:1 mixture of V. cholerae lacZ+ wild-type and V. cholerae lacZ- ΔbsrV is used to demonstrate the metabolic cooperation between producers and non-producers to outcompete other bacteria. - Incubate the co-cultivation chambers at 28 °C with mild agitation (100 rpm). Place the holes used for inoculation on the top to minimize sample spilling (Figure 1C). Samples from both compartments will be collected at different time points: 0, 24, 48 and 72 h.
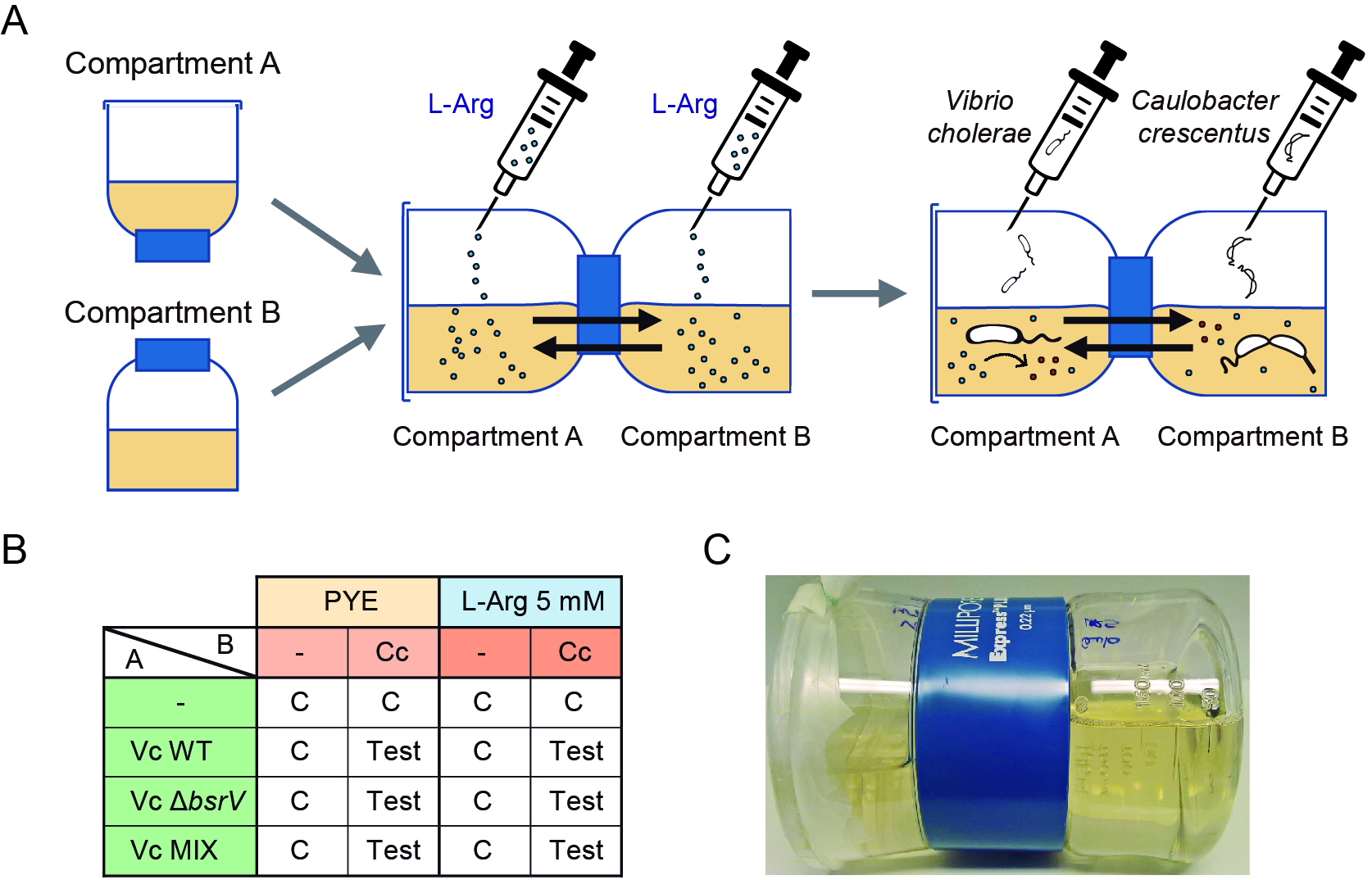
Figure 1. Preparation of the co-cultivation chambers. A. Fill both compartments of the Stericup filtration unit with the appropriate media. Attach and seal both parts and add the L-amino acid to a final concentration of 5 mM. Finally inoculate the compartments with the competing bacteria. B. Media and bacteria combinations for a complete competition experiment, including controls (C) and test samples (Test). Vc: V. cholerae. Cc: C. crescentus. C. Co-cultivation chamber. - At the desired time points, use 1 ml syringes and needles to collect 500 μl culture samples from each compartment and co-cultivation chamber and transfer to previously labeled microtubes.
- Viable cell count is performed immediately (see Procedure B). Transfer 200 μl of freshly collected samples from compartment A (V. cholerae) and B (C. crescentus) to a sterile 96-well plate.
- Determination of D-amino acid concentration is performed at the end of the whole experiment (see Procedure C). Centrifuge the remaining 300 μl sample at 21,000 x g for 5 min at room temperature using a microcentrifuge. Carefully transfer the supernatant (300 μl) to new microtubes and store samples at -20 °C until the end of the experiment. Make sure no cell pellet is transferred to the new tube.
- Streak V. cholerae cells from the freezer stock onto an LB agar plate and incubate upside-down for 16 h at 37 °C. V. cholerae lacZ+ wild-type and V. cholerae lacZ- ΔbsrV can be used to assess the role of BsrV in D-amino acid production.
- Viable cell count
Note: All steps need to be performed under sterile conditions, working by the flame or inside a laminar flow cabinet.- To determine the viable cell count of V. cholerae and C. crescentus, serially dilute cells 1:10. Transfer 20 μl from the previous dilution to a new well and add 180 μl of fresh medium (LB for V. cholerae, PYE for C. crescentus), thoroughly pipetting to make the mixture homogeneous. Following this procedure, dilute the cultures 9 times. Make sure to change the tips in every dilution step.
- Plate 100 μl of each dilution on agar plates (LB for V. cholerae, PYE for C. crescentus), for CFU count. First, add 6-10 sterile glass beads to each plate and then carefully pipette the bacterial culture. Then, close the lid and agitate the plates to homogeneously distribute the cell culture and let it dry. Remove the glass beads by carefully tilting the plates. Glass beads can be reused after decontamination, washing and sterilization. Incubate CFU count plates upside-down for 16 h at 37 °C for V. cholerae and 48 h at 28 °C for C. crescentus.
- Once grown, count colonies on the agar plates and calculate the viable CFU per ml based on the dilution factors applied (Figure 2).
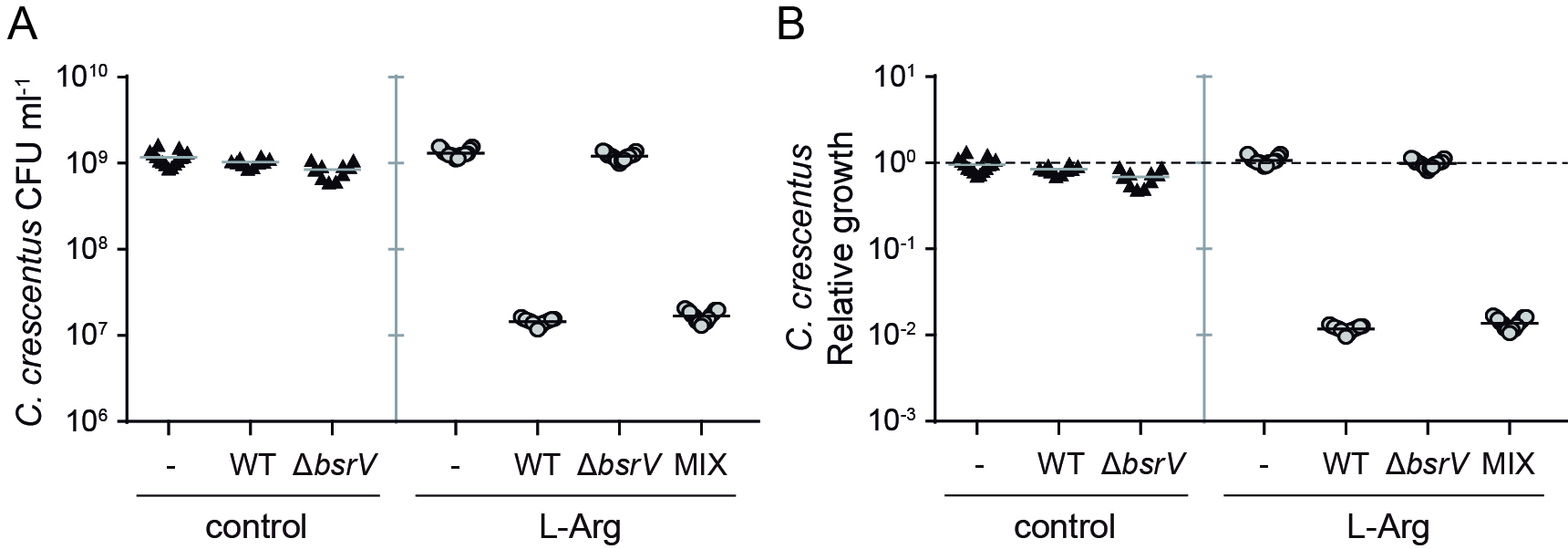
Figure 2. Viable cell count. A. C. crescentus growth expressed as CFU ml-1; B. Alternative representation using the relative growth compared to the control. -: C. crescentus control, WT: competition C. crescentus vs. V. cholerae lacZ+ wild-type, ΔbsrV: competition C. crescentus vs. V. cholerae lacZ- ΔbsrV, MIX: competition C. crescentus vs. a 1:1 mixture of V. cholerae lacZ+ wild-type and V. cholerae lacZ- ΔbsrV.
- To determine the viable cell count of V. cholerae and C. crescentus, serially dilute cells 1:10. Transfer 20 μl from the previous dilution to a new well and add 180 μl of fresh medium (LB for V. cholerae, PYE for C. crescentus), thoroughly pipetting to make the mixture homogeneous. Following this procedure, dilute the cultures 9 times. Make sure to change the tips in every dilution step.
- Determination of total D-amino acid concentration: DAAO assay
Note: No sterile conditions are required. In this two-step assay, DAAO produces α-ketoacid, NH3 and H2O2 from D-amino acids; peroxidase reduces H2O2 releasing free O2 that reacts with OPD, leading to the production of 2,3-diaminophenazine, a colorimetric product that can be detected using a spectrophotometer (Alvarez et al., 2018; Espaillat et al., 2014).- D-amino acid standard curve: prepare D-arginine dilutions at 0.05, 0.1, 0.25, 0.5, 1, 1.5 and 2 mM concentration in MilliQ water. Transfer 20 μl of each dilution to a clear flat-bottom 96-well plate. Transfer 20 µl of MilliQ water to another well to be used as a blank. For reproducibility, prepare triplicate standard curves and blanks.
- Supernatant samples: completely thaw the supernatant samples kept at -20 °C (let them stand on ice for 20 min). Prepare 1:2, 1:5 and 1:10 dilutions in new microtubes using MilliQ water. Transfer 20 μl of each sample (non-diluted, 1:2, 1:5 and 1:10) to the clear flat-bottom 96-well plate. For reproducibility, prepare triplicate reactions.
- Prepare the DAAO reaction buffer (see Recipes) and add 60 μl to each well using a multichannel pipette. Final reaction volume will be 80 μl.
- Close the 96-well plate lid to avoid evaporation and incubate for 1 h at 37 °C with vigorous shaking (400 rpm). Positive reactions will turn yellow.
- Using a multichannel pipette, add 2 volumes (160 μl) of 2 N HCl to each well to inactivate the reaction. The yellow color will turn orange.
- Read the absorbance at 492 nm using a microplate reader.
- D-amino acid standard curve: prepare D-arginine dilutions at 0.05, 0.1, 0.25, 0.5, 1, 1.5 and 2 mM concentration in MilliQ water. Transfer 20 μl of each dilution to a clear flat-bottom 96-well plate. Transfer 20 µl of MilliQ water to another well to be used as a blank. For reproducibility, prepare triplicate standard curves and blanks.
Data analysis
For reproducibility, biological samples should be tested in triplicate.
For testing cell viability, count the number of colonies grown on the plates and calculate the viable CFU per ml based on the dilution factors applied and the volume of culture plated (100 µl) (Figure 2).
Example: 153 colonies of C. crescentus in the plate with dilution 4 (1:104)
153 x 104 (dilution factor) x 10 (volume correction) = 1.53 x 107 CFU ml-1
The relative growth can be calculated by dividing the CFU ml-1 of every sample and condition by the CFU ml-1 in the control without L-amino acid or competitor bacteria.
To determine the total concentration of D-amino acid:
- Absorbance from the standard curve samples should fit to a linear regression model (Figure 3A). The goodness of the fit is represented by the R2 value.
- Use the equation of the linear regression model to calculate the total D-amino acid concentration of every sample and replica. Consider the dilution factors, if applied.
Discard all measurements with absorbance values above 1.2 units: above this value the model loses linearity and the extrapolation of the D-amino acid concentration is wrong. Use the values of the 1:2, 1:5 or 1:10 dilutions instead. - Total D-amino acid concentration can be represented as in Figure 3B. If needed, the basal D-amino acid concentration in PYE medium can be subtracted from the sample values.
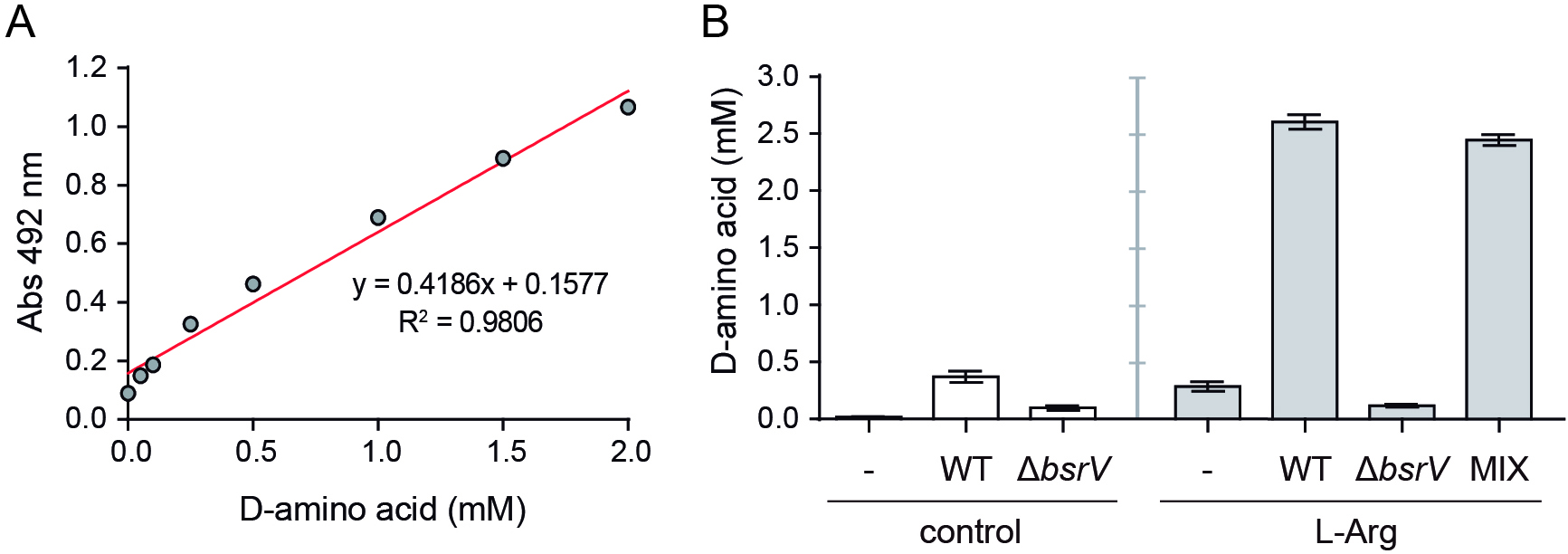
Figure 3. Determination of the D-amino acid concentration. A. Representative D-arginine standard curve; B. Total D-amino acid concentration in the media from co-cultivation chambers at 48 h. -: C. crescentus control, WT: competition C. crescentus vs. V. cholerae lacZ+ wild-type, ΔbsrV: competition C. crescentus vs. V. cholerae lacZ- ΔbsrV, MIX: competition C. crescentus vs. a 1:1 mixture of V. cholerae lacZ+ wild-type and V. cholerae lacZ- ΔbsrV.
Recipes
- LB medium
10.0 g L-1 tryptone
5.0 g L-1 yeast extract
10.0 g L-1 NaCl
Dissolve components in distilled water
Adjust the pH to 7.0 using 2 N NaOH
Adjust the final volume and sterilize by autoclaving (15 min at 121 °C and 1 atm)
Store at room temperature - PYE medium
2 g L-1 peptone
1.0 g L-1 yeast extract
1 ml L-1 1 M MgSO4
0.5 ml L-1 1 M CaCl2
Dissolve components in distilled water
Adjust the final volume and sterilize by autoclaving (15 min at 121 °C and 1 atm)
Store at room temperature - Agar plates
- For preparation of agar plates, dissolve the medium components in distilled water, add 15 g L-1 bacteriological agar, then adjust pH if needed and the final volume and finally sterilize by autoclaving (15 min at 121 °C and 1 atm)
- Let the medium cool down to 50 °C and pour in sterile Petri dishes under sterile conditions (approximately 20 ml per plate). Let the plates solidify at room temperature. Store plates at 4 °C
- For preparation of agar plates, dissolve the medium components in distilled water, add 15 g L-1 bacteriological agar, then adjust pH if needed and the final volume and finally sterilize by autoclaving (15 min at 121 °C and 1 atm)
- L- and D-amino acid stock solutions
Dissolve the corresponding amount of L- or D-arginine in MilliQ water to a final concentration of 200 mM
Sterilize using 0.22 µm pore size filter units
Store at room temperature - Sodium phosphate buffer 500 mM pH 7.5
12.9 g L-1 NaH2PO4·H2O
109.1 g L-1 Na2HPO4·7H2O
Dissolve components in distilled water
Adjust the pH to 7.5 using ortho-phosphoric acid 25% (v/v) or 2 N NaOH, and adjust the final volume
Store at room temperature - DAAO reaction buffer
Per reaction, prepare 60 µl final volume buffer (see table below) containing sodium phosphate buffer 33.3 mM pH 7.5, FAD 8.3 µg ml-1, freshly prepared OPD 83.3 µg ml-1, horseradish peroxidase 41.7 µg ml-1 and Trigonopsis variabilis DAAO (Komarova et al., 2012) 33.3 µg ml-1.
All stock solutions are prepared in MilliQ water unless otherwise specified. Aliquot and store FAD, horseradish peroxidase and DAAO stock solutions at -20 °C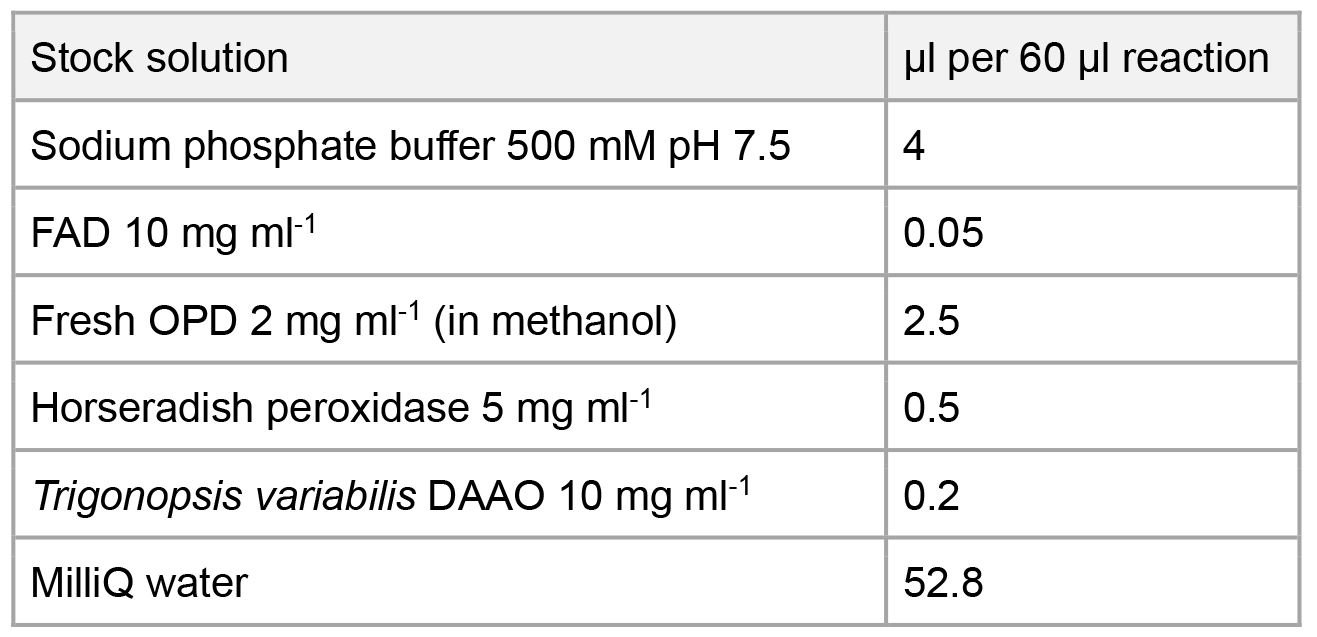
Acknowledgments
This work reports in detail the bacterial competition assay previously used to demonstrate the use of D-arginine by V. cholerae to outcompete other bacteria (Alvarez et al., 2018). This work was funded by The Knut and Alice Wallenberg Foundation (KAW), The Laboratory of Molecular Infection Medicine Sweden (MIMS), the Swedish Research Council and the Kempe Foundation. The authors declare no conflict of interest or competing interest.
References
- Alvarez, L., Aliashkevich, A., de Pedro, M. A. and Cava, F. (2018). Bacterial secretion of D-arginine controls environmental microbial biodiversity. ISME J 12(2): 438-450.
- Bopp, M. (1965). [Inhibition of Agrobacterium tumefaciens by D-amino acids]. Z Naturforsch B 20(9): 899-905.
- Caparros, M., Pisabarro, A. G. and de Pedro, M. A. (1992). Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol 174(17): 5549-5559.
- Cava, F., de Pedro, M. A., Lam, H., Davis, B. M. and Waldor, M. K. (2011). Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J 30(16): 3442-3453.
- Espaillat, A., Carrasco-Lopez, C., Bernardo-Garcia, N., Pietrosemoli, N., Otero, L. H., Alvarez, L., de Pedro, M. A., Pazos, F., Davis, B. M., Waldor, M. K., Hermoso, J. A. and Cava, F. (2014). Structural basis for the broad specificity of a new family of amino-acid racemases. Acta Crystallogr D Biol Crystallogr 70(Pt 1): 79-90.
- Fox, S., Fling, M. and Bollenback, N. (1944). Inhibition of bacterial growth by D-leucine. J Biol Chem 155: 465-468.
- Grula, E. A. (1960). Cell division in a species of Erwinia. I. Inhibition of division by D-amino acids. J Bacteriol 80: 375-385.
- Kobayashi, Y., Fling, M. and Fox, S. W. (1948). Antipodal specificity in the inhibition of growth of Escherichia coli by amino acids. J Biol Chem 174(2): 391-398.
- Komarova, N. V., Golubev, I. V., Khoronenkova, S. V., Chubar, T. A. and Tishkov, V. I. (2012). Engineering of substrate specificity of D-amino acid oxidase from the yeast Trigonopsis variabilis: directed mutagenesis of Phe258 residue. Biochemistry (Mosc) 77(10): 1181-1189.
- Lam, H., Oh, D. C., Cava, F., Takacs, C. N., Clardy, J., de Pedro, M. A. and Waldor, M. K. (2009). D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325(5947): 1552-1555.
- Lark, C. and Lark, K. G. (1959). The effects of D-amino acids on Alcaligenes fecalis. Can J Microbiol 5: 369-379.
- Tuttle, A. L. and Gest, H. (1960). Induction of morphological aberrations in Rhodospirillum rubrum by D-amino acids. J Bacteriol 79: 213-216.
- Yaw, K. E. and Kakavas, J. C. (1952). Studies on the effects of D-Amino acids on Brucella abortus. J Bacteriol 63(2): 263-268.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Alvarez, L. and Cava, F. (2018). Bacterial Competition Assay Based on Extracellular D-amino Acid Production. Bio-protocol 8(7): e2787. DOI: 10.21769/BioProtoc.2787.
Category
Microbiology > Microbial physiology > Interspecific competition
Microbiology > Microbial signaling > Interspecies communication
Biochemistry > Other compound > Amino acid > D-amino acids
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









