- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ciliary Assembly/Disassembly Assay in Non-transformed Cell Lines
(*contributed equally to this work) Published: Vol 8, Iss 6, Mar 20, 2018 DOI: 10.21769/BioProtoc.2773 Views: 10366
Reviewed by: Ralph Thomas BoettcherChris TibbittMurugappan Sathappa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Embryonic Cardiomyocytes and Cell Proliferation Assay Using Genetically Engineered Reporter Mouse Model
Maren Beall [...] Jihyun Jang
Sep 5, 2023 1777 Views

Flow-based In Vivo Method to Enumerate Translating Ribosomes and Translation Elongation Rate
Mina O. Seedhom [...] Jonathan W. Yewdell
Jan 20, 2025 3042 Views
Abstract
The primary cilium is a non-motile sensory organelle whose assembly and disassembly are closely associated with cell cycle progression. The primary cilium is elongated from the basal body in quiescent cells and is resorbed as the cells re-enter the cell cycle. Dysregulation of ciliary dynamics has been linked with ciliopathies and other human diseases. The in vitro serum-stimulated ciliary assembly/disassembly assay has gained popularity in addressing the functions of the protein-of-interest in ciliary dynamics. Here, we describe a well-tested protocol for transfecting human retinal pigment epithelial cells (RPE-1) and performing ciliary assembly/disassembly assays on the transfected cells.
Keywords: Primary ciliumBackground
Primary cilia are hair-like sensory organelles that appear at the G0/G1 phase, and are disassembled prior to the S phase of the cell cycle (Tucker et al., 1979). Previous studies have confirmed that certain non-transformed cell types (i.e., RPE-1 cells, 3T3 fibroblasts, and mouse embryonic fibroblasts [MEFs]) can be starved to induce quiescence and ciliary formation. Subsequent re-addition of serum triggers biphasic ciliary resorption, which peaks at 2 h and 24 h following stimulation (Tucker et al., 1979; Li et al., 2011). This phenomenon lays the foundation for the serum-stimulated ciliary assembly/disassembly assay commonly used in the literature to identify proteins involved in ciliary assembly and disassembly (Pugacheva et al., 2007; Saito et al., 2017). MEFs derived from transgenic mice (in which the gene-of-interest is deleted) are often used to investigate the dynamic role of a given protein in the ciliary assembly/disassembly assays. When the specific MEF types are not accessible, one may modify the expression level of the targeted protein in naive RPE-1 cells (or 3T3 or MEFs) using transfection of cDNA or short hairpin RNA. We describe the protocol of these procedures that are routinely carried out in the lab. To unambiguously identify the cell autonomous effect on ciliary assembly or disassembly in the transfected cells, we typically ‘tag’ the transfected cells with green fluorescence via the expressed GFP or GFP-fusion protein.
Materials and Reagents
- 10 µl pipette tips (Corning, catalog number: 4110 , or Denville Scientific, catalog number: P1096 )
- 200 µl pipette tips (FUKAEKASEI and WATSON, catalog number: 110-705Y , or Denville Scientific, catalog number: P1122 )
- 1,000 µl pipette tips (FUKAEKASEI and WATSON, catalog number: 110-706B , or Denville Scientific, catalog number: P2103-N )
- 100 mm cell culture dish (AS ONE, Violamo, catalog number: 2-8590-03 , or Corning, catalog number: 430167 )
- 1.5 ml microcentrifuge tube (Sorenson BioScience, catalog number: 11510 , or National Scientific, catalog number: CN1700-BP )
- Precleaned, sterile 12-13 mm micro-glass coverslips (Matsunami Glass, catalog number: C013001 , or Thermo Fisher Scientific, catalog number: 12CIR-1.5 )
- 35 mm cell culture dish (Corning, Falcon®, catalog number: 353001 , or Corning, catalog number: 430165 )
- 15 ml centrifuge tube (FUKAEKASEI and WATSON, catalog number: 1332-015S , or Corning, catalog number: 430053 )
- 24-well plate (NuncTM, Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 142475 , or Corning, catalog number: 3527 )
- Parafilm
- 14 x 16 cm glass (or plastic) plates
- Micro-slide glass (Matsunami Glass, catalog number: S024410 )
- 0.22 µm filter (Corning, catalog number: 431097 , or Advantec MFS, catalog number: 25CS020AS )
- RPE-1 cells (ATCC, catalog number: CRL-4000 )
- Midi-prepared, or maxi-prepared plasmid (> 1 µg/µl is preferred)
Note: We typically have the cDNA (under the CAG or CMV promoter) or short hairpin RNA (shRNA; under the U6 promoter) inserted in the same plasmid that also expresses GFP (e.g., pCAGIG vector). - 0.05% Trypsin-0.53 mM EDTA (Wako Pure Chemical Industries, catalog number: 202-16931 )
- Fetal bovine serum (FBS) (PAA Laboratories, catalog number: A15-701 )
- Dulbecco’s modified Eagle medium/Ham’s F-12 (DMEM/F12) (Wako Pure Chemical Industries, catalog number: 048-29785 )
- Methanol
- Monoclonal anti-acetylated α-tubulin (Ac-Tub) antibody, clone 6-11B-1 (Sigma-Aldrich, catalog number: T6793 )
- Anti-Tubulin Antibody, Detyrosinated (Merck, catalog number: AB3201 )
- Monoclonal anti-γ-tubulin antibody, clone GTU-88 (Sigma-Aldrich, catalog number: T5326 )
- Polyclonal anti-GFP tag antibody (Thermo Fisher Scientific, InvitrogenTM, catalog number: A-6455 )
- Goat anti-mouse IgG (H+L) antibody, Alexa Fluor 568 (Thermo Fisher Scientific, InvitrogenTM, catalog number: A-11004 )
- Goat anti-mouse IgG2b antibody, Alexa Fluor 568 (Thermo Fisher Scientific, InvitrogenTM, catalog number: A-21144 )
- Goat anti-mouse IgG1 antibody, Alexa Fluor 647 (Thermo Fisher Scientific, InvitrogenTM, catalog number: A-21240 )
- Goat anti-Rabbit IgG (H+L) antibody, Alexa Fluor 488 (Thermo Fisher Scientific, InvitrogenTM, catalog number: R37116 )
- Fluorescent mounting medium (Agilent Technologies, Dako, catalog number: S302380-2 , code number: S3023) or ProLong Gold Antifade Mountant (Thermo Fisher Scientific, InvitrogenTM, catalog number: P36930 )
- Nail polish
- Dulbecco’s modified Eagle medium (DMEM) (NACALAI TESQUE, catalog number: 08458-16 )
- 100 mM pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 11360070 )
- 10x D-PBS (-) (Wako Pure Chemical Industries, catalog number: 048-29805 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Wako Pure Chemical Industries, catalog number: 031-00435 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Wako Pure Chemical Industries, catalog number: 135-00165 )
- 16% Paraformaldehyde (PFA) aqueous solution (Electron Microscopy Science, catalog number: 15710 )
- Ammonium chloride (NH4Cl) (Wako Pure Chemical Industries, catalog number: 017-02995 )
- Bovine serum albumin (BSA) (Wako Pure Chemical Industries, catalog number: 010-15114 )
- Triton X-100 (GE Healthcare, catalog number: 17-1315-01 )
- Sodium azide (NaN3) (Wako Pure Chemical Industries, catalog number: 190-01272 )
- 4’,6-Diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D1306 )
- Growth medium (see Recipes)
- PBS for cell culture (see Recipes)
- 2x PBSc/m (see Recipes)
- PBSc/m (see Recipes)
- 4% PFA/PBSc/m (see Recipes)
- 50 mM NH4Cl (see Recipes)
- BTPAD (see Recipes)
Equipment
- 5% CO2/95% air tissue culture incubator (SANYO, model: MCO-18AIC )
- Water bath (TOKYO RIKAKIKAI, Eyela, model: NTT-1200 )
- Hemocytometer chamber (Erma, catalog number: 03-303-1 )
- Low-speed centrifuge (TOMY SEIKO, model: LC-100 ; Eppendorf, model: 5702 )
- Pipette 20 µl (Pipetman P) (Gilson, catalog number: F123600 )
- Pipettes 200 µl (Pipetman P) (Gilson, catalog number: F123601 )
- Pipettes 1,000 µl (Pipetman P) (Gilson, catalog number: F123602 )
- For Procedure B1:
NeonTM transfection system (Thermo Fisher Scientific, InvitrogenTM, catalog number: MPK5000 ) - For Procedure B2:
- NucleofectorTM device (Lonza, model: NucleofectorTM I )
- Cuvettes PlusTM Electroporation Cuvettes & transfer tips (BTX, catalog number: 45-0135 )
- Amaxa® Cell Line Nucleofector® Kit V (Lonza, catalog number: VCA-1003 )
- NucleofectorTM device (Lonza, model: NucleofectorTM I )
- Forceps (Fine Scientific Tool, Dumont, model: #5/45 )
- Autoclave (TOMY SEIKO, model: LBS-245 )
- Epifluorescent microscope (Carl Zeiss, model: Axioplan 2 imaging ) equipped with objective lens (EC Plan-Neofluar 40x/0.75, Carl Zeiss, catalog number: 440350-9903-000 )
- Confocal microscope (ZEISS, model: LSM-780 )
Procedure
The present assay is performed by the following timeline and procedures (Figure 1). In the present protocol, we show the preparation of four samples for each transfection (e.g., before assembly, 0 h-, 2 h- and 24 h-after serum re-addition). Each researcher can prepare samples depending on the need. For example, when we focus on mechanisms of ciliary disassembly, we typically prepare three samples for each transfection (0 h-, 2 h- and 24 h-after serum re-addition). Additional sample(s) can be added for testing the transfection efficiency (on Day 3).
Day 1: Cell subculture (see Procedure A)
Day 2: Cell transfection (see Procedure B); Induction of ciliary assembly (see Procedure C)
Day 3: Checking cell transfection efficiency
Day 4: Induction of ciliary disassembly (see Procedure D); Early time point (e.g., 0 h, 2 h) cell harvest (see Procedure E)
Day 5: Late time point (e.g., 24 h) cell harvest
Immunostaining (see Procedure F) and data analysis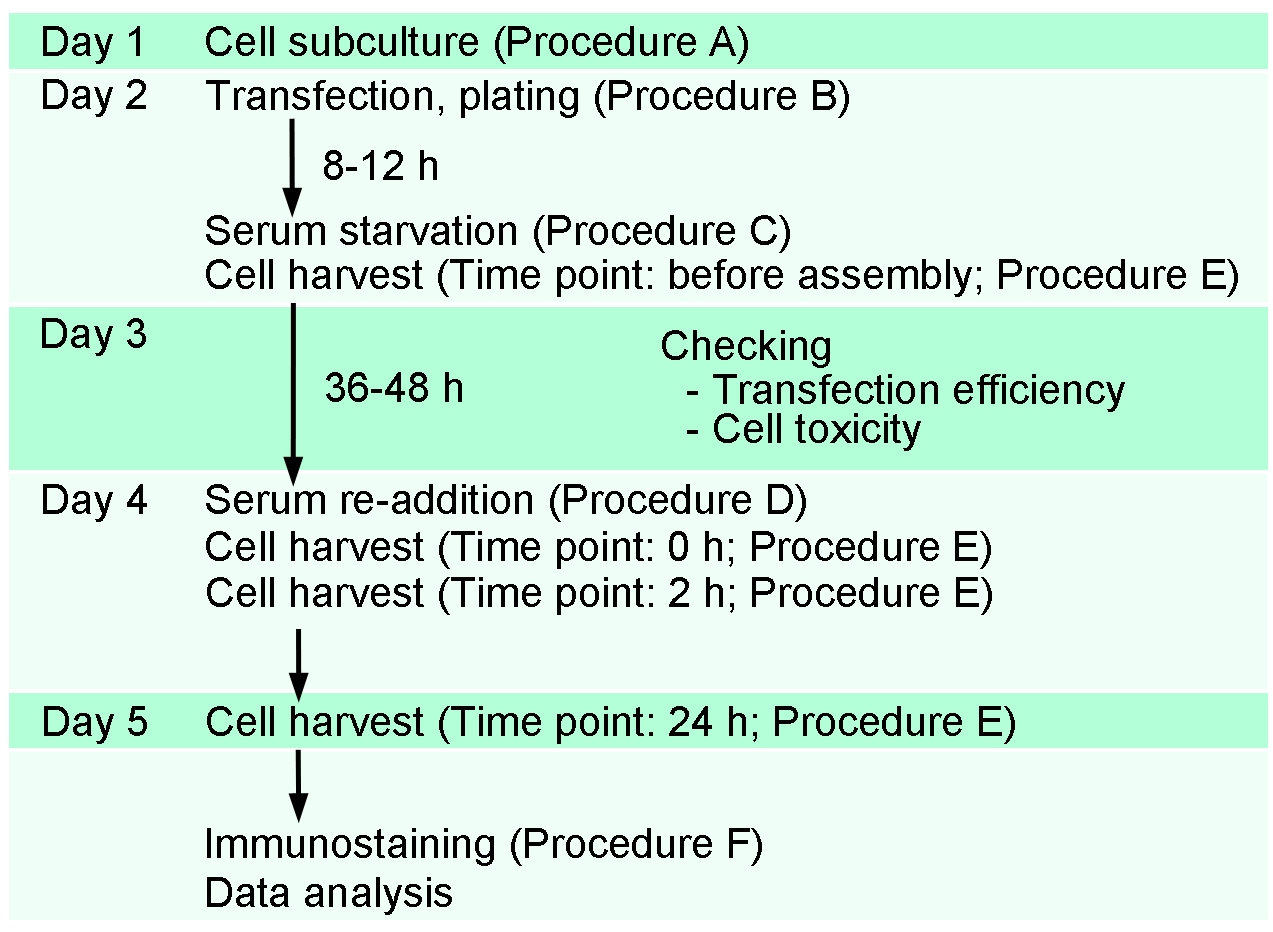
Figure 1. A schematic representation of the timeline of transfection and ciliary assembly/disassembly assay
- Cell subculture (Day 1)
RPE-1 cells maintained in 10% FBS-containing growth medium (see Recipes) in a 5% CO2 humidified incubator at 37 °C are passaged and plated on 100 mm cell culture dishes using standard techniques. The cells are plated at a density expecting 50-80% confluence on the second day. In general, one 100 mm dish of cells (total of ~2-4 x 106 cells) is sufficient for 4-6 transfection reactions when Procedure B1 is used (3 x 105 cells for each reaction) and 2-4 transfection reactions when Procedure B2 is used (1 x 106 cells for each reaction).
Note: We avoid using old (> 15 passage) or overgrown RPE-1 cells, which tend to have a poor cilium induction rate. - Cell transfection (Day 2)
NeonTM (Procedure B1) and NucleofectorTM (Procedure B2) transfection systems generate similar transfection efficiencies and low cell death. More than 60% of GFP plasmid-transfected RPE-1 cells express green fluorescence on the second day after transfection. In contrast, Lipofectamine 2000® or polyethylenimine (PEI) do not generate a high number of GFP-expressing cells.
Prepare the following before conducting the transfection procedures described in Procedure B1 or B2.- Prewarm growth medium in a 37 °C water bath. Place a single plasmid (or premixed multiple plasmids, when applicable) in a sterile 1.5 ml tube. For the transfection reactions of Procedures B1 and B2, ~3 µg and ~6 µg of DNA are to be used, respectively.
If performing Procedure B2, prepare the Amaxa transfection master mixture. For each transfection reaction, mix 85.9 µl of solution I and 19.1 µl of solution II by gentle pipetting and quick spin. Prepare excess reactions to account for pipetting error. - Place three or more acid-washed sterile coverslips in each 35 mm cell culture dish. Add 1 ml of growth medium and transfer to a 5% CO2 incubator at 37 °C.
- For each transfection reaction, prepare 1 ml of prewarmed growth medium in a 1.5 ml sterile tube and leave it in a 37 °C water bath.
- Use trypsin-EDTA solution to detach RPE-1 cells (0.5 ml per 100 mm plate). After ~2 min of incubation at 37 °C, collect the detached cells into a 15 ml centrifuge tube containing 9.5 ml of warmed growth medium, in which the FBS inactivates trypsin.
- Count the number of cells using a hemocytometer chamber.
B1. NeonTM transfection system- Centrifuge the cell suspension at 280 x g (1,200 rpm) for 2 min at room temperature.
- Aspirate the supernatant. Resuspend the cell pellet in 10 ml of sterile PBS (see Recipes).
- Centrifuge the cell suspension at 280 x g (1,200 rpm) for 2 min at room temperature.
- Carefully aspirate all of the supernatant. Resuspend every 1 x 106 cells with 100 µl of Resuspension Buffer R (included in NeonTM Transfection System). Thirty microliters of cell suspension (i.e., 3 x 105 cells) is required for each transfection.
- Gently mix 3 µg of the plasmid in 30 µl of cell suspension by pipetting.
- Use a 10 µl NeonTM Tip to obtain 10 µl of cell-DNA mixture and then apply an electrical pulse according to the manufacturer’s protocol using the following parameters (Pulse voltage ‘1,050 V’, Pulse width ‘35 msec’, and Pulse number ‘2 times’). Repeat until the entire volume (30 µl) of the cell-DNA mixture is ‘pulsed’.
- Immediately after each pulse, transfer the transfected cells into a 1.5 ml tube containing 1 ml of warmed growth medium. Use the same tube to collect all 3 x 105 transfected cells.
- Plate the transfected cells onto a 35 mm dish containing coverslips and immediately transfer to a 5% CO2 incubator at 37 °C for 12 h.
B2. NucleofectorTM transfection system- For each transfection reaction, transfer a volume of cell suspension equivalent to 1 x 106 trypsin-EDTA-detached RPE-1 cells into a 15 ml tube.
- Centrifuge the cell suspension at 280 x g (1,200 rpm) for 2-3 min at room temperature.
- Carefully aspirate all of the supernatant. Gently resuspend the cell pellet with 100 µl of Amaxa transfection master mixture using a P200 pipette.
Note: Up to 4 transfection reactions may be carried out in the same batch by pooling 4 x 106 cells in 400 µl of Amaxa transfection master mixture. - For each transfection reaction, transfer 100 µl of the cell suspension into a 1.5 ml tube containing DNA. Gently pipette no more than twice and avoid air bubbles.
- Transfer the entire cell-DNA mixture into an Electroporation Cuvette (BTX). Do not pipette out the last drop to avoid air bubbles.
- Place the cuvette in the NucleofectorTM device and pulse the cells with the program ‘T23’. This is a program preset in the device, which can be selected by a simple button pressing.
- Immediately after the pulse, collect the transfected cells in the cuvette using a fine tip dropper (included in the individually packaged Electroporation Cuvette). Transfer them into a 1.5 ml tube containing 1 ml of warmed growth medium.
- Plate the transfected cells onto the 35 mm dish containing coverslips and growth medium, and immediately transfer the cells to a 5% CO2 incubator at 37 °C for 12 h.
- Prewarm growth medium in a 37 °C water bath. Place a single plasmid (or premixed multiple plasmids, when applicable) in a sterile 1.5 ml tube. For the transfection reactions of Procedures B1 and B2, ~3 µg and ~6 µg of DNA are to be used, respectively.
- Induction of ciliary assembly (Day 2)
- Approximately 8-12 h after cell transfection (Procedure B), the cells should have settled at the bottom. Follow Procedure E in the case that researcher wishes harvest cell samples before serum starvation (e.g., observing cilia before serum starvation or testing the transfection efficiency by immunostaining). Remove the culture medium, gently rinse the cells in 2 ml of PBS (three times), and add 2 ml of pre-warmed serum-free DMEM (or DMEM/F12). We do not notice any obvious differences between DMEM and DMEM/F12 in this step for the assembly or the subsequent disassembly of cilia.
- Incubate in a 5% CO2 incubator at 37 °C for 36-48 h. Optional: during this period (on Day 3), we use an inverted phase/fluorescent microscope to check the green fluorescence-positive cells to estimate the transfection efficiency. We also pay attention to the cell toxicity by observing the number of the cells detached from the coverslips/dishes.
- Approximately 8-12 h after cell transfection (Procedure B), the cells should have settled at the bottom. Follow Procedure E in the case that researcher wishes harvest cell samples before serum starvation (e.g., observing cilia before serum starvation or testing the transfection efficiency by immunostaining). Remove the culture medium, gently rinse the cells in 2 ml of PBS (three times), and add 2 ml of pre-warmed serum-free DMEM (or DMEM/F12). We do not notice any obvious differences between DMEM and DMEM/F12 in this step for the assembly or the subsequent disassembly of cilia.
- Induction of ciliary disassembly (Day 4)
- At the end of serum-starvation, using a pair of sterile forceps, remove one coverslip and immediately wash and fix them (0 h time-point; see Procedure E). Aspirate the serum-free medium and replace it with growth medium.
- Incubate the cells in a 5% CO2 incubator at 37 °C for the indicated periods; 2 h and 24 h would be good starting points.
- At the end of serum-starvation, using a pair of sterile forceps, remove one coverslip and immediately wash and fix them (0 h time-point; see Procedure E). Aspirate the serum-free medium and replace it with growth medium.
- Cell harvest (Day 2, 4 and 5)
- Transfer one or more coverslips from the incubated 35 mm dish into an empty 24-well plate using a pair of sterile forceps with the cell-side facing up.
- Rinse the coverslip in 1 ml of PBSc/m (see Recipes) three times and fix in 0.5 ml of 4% PFA/PBSc/m (see Recipes) for 10 min at room temperature.
- Rinse the coverslip in 1 ml of PBSc/m once, and fill with 1 ml of another PBSc/m.
- Transfer one or more coverslips from the incubated 35 mm dish into an empty 24-well plate using a pair of sterile forceps with the cell-side facing up.
- Immunostaining
Primary cilium can be visualized by immunostaining acetylated α-tubulin (Ac-Tub) or detyrosinated tubulin (Figure 2). Basal body can be detected by γ-tubulin (γ-Tub) immunostaining (Figure 2B). The γ-Tub staining conveniently indicates the proximal end of the cilium. It is also a good landmark particularly in the cells with a short or no detectable cilium. However, to obtain γ-Tub stained signal, the cells have to be treated with cold methanol (see Step F2).
Each researcher can perform immunostaining procedure according to own preference, such as the procedures described previously (Phua et al., 2017; Shnitsar et al., 2015; Tomoshige et al., 2017). Below, we describe our procedures of triple staining for Ac-Tub, γ-Tub and GFP. GFP staining is used to enhance the detection of transfected cells that express GFP (or GFP-fusion proteins). So that role of the target molecule(s) in ciliary assembly/disassembly can be unambiguously identified in GFP-positive transfected cells.
The entire procedure detailed below is performed in a light-blocking humidity chamber (homemade) at room temperature.- Place a piece of Parafilm on 14 x 16 cm of glass (or plastic) plates in the chamber.
- Pre-treat the previously fixed coverslip with pre-chilled methanol for 1-3 min at -20 °C.
- Immediately rinse the cells with PBSc/m, two times.
- Place the coverslip on the Parafilm with the cell-side facing up.
- Add ~300 µl of 50 mM NH4Cl (see Recipes) from the edge of the coverslips using a P1000 pipette (or a dropper) and incubate for 10 min.
- Aspirate the NH4Cl from the edge of the coverslips, and rinse in 300 µl of PBSc/m once.
- Incubate in 100 µl of BTPAD (see Recipes) for 30 min.
- Incubate in 50-100 µl of primary antibody-containing BTPAD for 60 min at room temperature. Antibody dilution: anti-Ac-Tub (mouse IgG2b, 1:1,000), anti-γ-Tub (mouse IgG1, 1:1,000), anti-GFP (rabbit IgG, 1:1,000).
- Remove unbound primary antibodies by incubating the cells in 300 µl of PBSc/m for 5 min, three times.
- Incubate in 50-100 µl of secondary antibody-containing BTPAD for 45 min at room temperature. Antibody dilution: Alexa Fluor 568-conjugated goat anti-mouse IgG2b (for anti-Ac-Tub, 1:400), Alexa Fluor 647-conjugated goat anti-mouse IgG1 (for anti-γ-Tub, 1:400), and Alexa Fluor 488-conjugated goat anti-rabbit IgG (for anti-GFP, 1:400).
Note: When the cells are labeled for Ac-Tub, but not for γ-Tub, in Step F8, the isotype specific secondary antibody is not necessary. We use Alexa Fluor 568-conjugated anti-mouse IgG (H+L) antibody instead. - Rinse in 300 µl of PBSc/m for 5 min, three times.
- Remove the excess PBSc/m, and wipe the side of coverslips without cells.
- Mount the coverslip with 4 µl of mounting media on a micro-slide glass.
- Seal the rim of coverslip with clear nail polish.
- Observe samples under an epifluorescence microscope (ZEISS, Axioplan 2 imaging) or a confocal microscope (ZEISS, model: LSM-780). For an epifluorescence microscope, objective lens: 40x magnification without immersion. Ocular lens: 10x magnification. For a confocal microscope, objective lens: 63x magnification, oil immersion. Representative images taken on the confocal microscopy are shown in Figure 2.
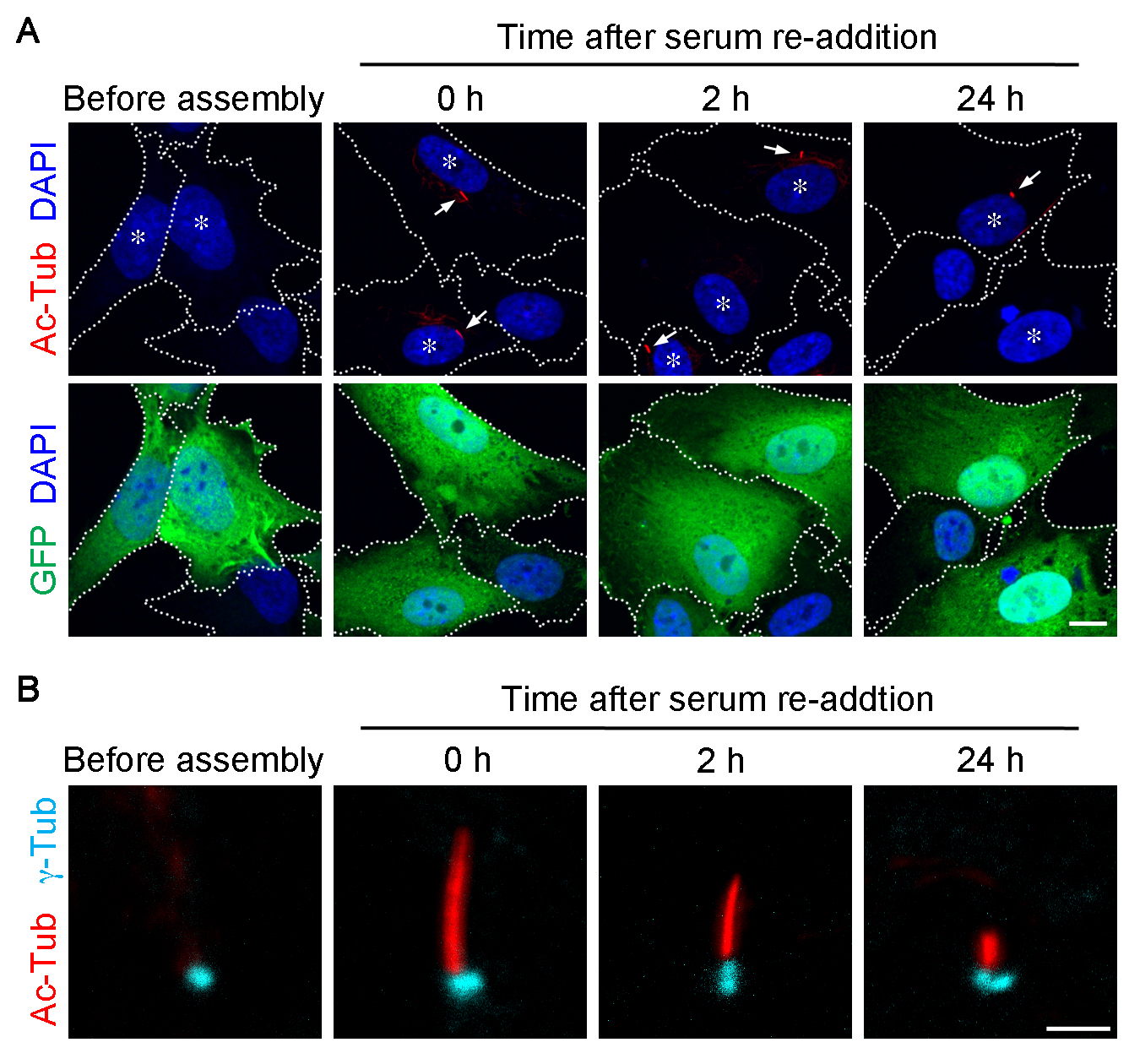
Figure 2. Primary cilia are assembled and disassembled by depletion and re-addition of serum, respectively. Representative images of primary cilia assembled in RPE-1 cells. For presentation purpose, images were acquired using a confocal microscope (ZEISS, LSM-780). The cells were harvested before serum starvation (before assembly), 36 h after the serum-starvation (0 h), and the subsequent serum re-addition for 2 h and 24 h, and labeled for Ac-Tub (red; A, B), GFP (green; A), and γ-Tub (cyan; B). Nucleus was stained with DAPI (blue; A). Dashed lines demarcate the cell borders. Asterisks in (A) highlight the GFP+ cells. Arrows in (A) point to the cilia in GFP+ cells. Scale bars = 10 µm (A) and 2 µm (B).
- Place a piece of Parafilm on 14 x 16 cm of glass (or plastic) plates in the chamber.
Data analysis
Because the length of the cilium is positively correlated with the number of cells exhibiting cilia, and because measuring the cilium length is labor-intensive, the number of cells exhibiting detectable cilium is counted as a surrogate index instead (Li et al., 2011).
Notes: For early-stage researchers, we recommend observing cilia with higher-magnification lens (e.g., 63x, 100x). However, we must note that researchers need to adjust Z-positions to find each cilium between cells of interest due to the higher magnification.
- Count the number of cells expressing Ac-Tub among GFP+-transfected cells in a double-blind fashion. More than 100 cells are counted in each experiment (two sets of 50 cells; one from the left side and the other from the right side of each coverslip). At least three independent experiments are performed for each condition. Data from independent experiments are represented as the mean percentages ± SEM. One-way analysis of variance (ANOVA) or two-way ANOVA is followed by Tukey’s test or Bonferroni’s test (as a post hoc test), respectively (Figure 3).
- Figure 3A shows a time-dependency of ciliary assembly after serum removal in native RPE-1 cells. Eighty percent of cells expressed cilia 36 h after the serum-starvation.
- Figure 3B shows the representative results of ciliary disassembly assay. An example of ciliary disassembly by knockdown of Tctex-1 is also shown. Tctex-1 is a light chain of cytoplasmic dynein complex and is an indispensable molecule for ciliary disassembly (Li et al., 2011; Yeh et al., 2013; Saito et al., 2017). Both U6 promoter and short hairpin RNA (shRNA) sequence of Tctex-1 were inserted into a pCAGIG vector that also encoded GFP (see Materials and Reagents #13) (Li et al., 2011). Cells transfected with vector (Control) or Tctex-1-shRNA (Tctex-1-sh) were serum-starved for 36 h and retreated with growth medium for 2 h and 24 h. The number of cells with cilia was counted among GFP+-transfected cells. Percentage of cells with cilia decreased from 80% (0 h) to 60% (2 h) and 40% (24 h) after serum re-addition in the cells of the control setup. Although the ciliary assembly was unaffected in Tctex-1-shRNA transfected cells, the rate of ciliary disassembly in these cells was significantly suppressed.
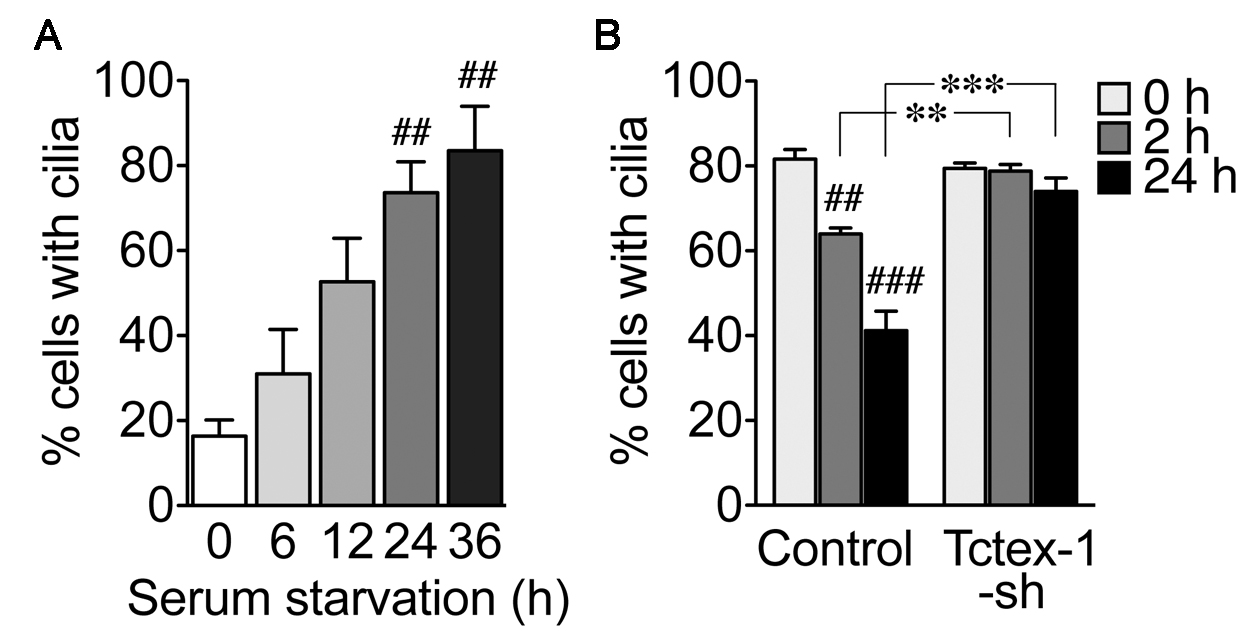
Figure 3. Representative results from ciliary assembly and disassembly assays. A. Ciliary assembly assay. Naive RPE-1 cells were harvested at indicated time points after serum starvation. Y-axis showed the percentage of the cells expressing cilia. B. Ciliary disassembly assay. RPE-1 cells, transfected with control vector or Tctex-1-shRNA that also expressed GFP, were first starved and then treated with grown medium for the indicated time periods. Y-axis showed the percentage of the GFP+ cells with cilia. ##P < 0.01, ###P < 0.001; one-way ANOVA followed by Tukey’s test (comparing to the 0 h time point of each group). **P < 0.01, ***P < 0.001; two-way ANOVA followed by Bonferroni’s test (comparing between groups). n = 100 cells per experiment, three (A) and five (B) independent experiments.
Recipes
- Growth medium
- Take 500 ml of DMEM/F12
- Add 50 ml of FBS (final 10%)
- Add 5.5 ml of 100 mM pyruvate (final 1 mM)
- Store at 4 °C
- Take 500 ml of DMEM/F12
- PBS for cell culture
- Take 180 ml of ddH2O
- Add 20 ml of 10x D-PBS (-)
- Autoclave
- Store at room temperature
- Take 180 ml of ddH2O
- 2x PBSc/m
- Take 795 ml of ddH2O
- Add 200 ml of 10x D-PBS (-)
- Add 400 µl of 1 M CaCl2 (final 0.4 mM)
- Add 4,000 µl of 1 M MgCl2 (final 4 mM)
- Filter through a 0.22 µm filter
- Store at room temperature
- Take 795 ml of ddH2O
- PBSc/m
- Take 500 ml of ddH2O
- Add 500 ml of 2x PBSc/m
- Filter through a 0.22 µm filter
- Store at room temperature
- Take 500 ml of ddH2O
- 4% PFA/PBSc/m
Prepare just before the procedure in a chemical hood- Take 1 ml of ddH2O
- Add 2 ml of 2x PBSc/m
- Add 1 ml of 16% PFA
- Take 1 ml of ddH2O
- 50 mM NH4Cl
- Take 20 ml of ddH2O
- Add 25 ml of 2x PBSc/m
- Add 133.75 mg of NH4Cl (final 50 mM)
- Dilute up to 50 ml with ddH2O
- Store at room temperature
- Take 20 ml of ddH2O
- BTPAD
- Take 25 ml of PBSc/m
- Add 625 µl of 20% BSA (final 0.5%)
- Add 313 µl of 20% Triton X-100 (final 0.25%)
- Add 250 µl of 2% NaN3 (final 0.02%)
- Add 2.5 µl of 3 mM DAPI (final 0.3 µM)
- Filter through a 0.22 µm filter
- Store at 4 °C
- Take 25 ml of PBSc/m
Acknowledgments
This protocol was adapted from a previously published study (Saito et al., 2017). This work was supported by NIH RO1 EY11307, EY016805, Research To Prevent Blindness, Stem Innovation Award recipient (RPB), and Kaohsiung Medical University Research Foundation (105KMUOR02) (to C.-H. S.); Grant-in-Aid for Scientific Research from the Japan Society for Promotion of Science (No. 23770136 and No. 15K20856 to M.S.) and Takeda Science Foundation (to M.S.). We would like to acknowledge the Biomedical Research Core (Tohoku University Graduate School of Medicine). We would also like to thank Editage (www.editage.jp/) for English language editing. The authors declare no competing financial interests.
References
- Li, A., Saito, M., Chuang, J. Z., Tseng, Y. Y., Dedesma, C., Tomizawa, K., Kaitsuka, T. and Sung, C. H. (2011). Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol 13(4): 402-411.
- Phua, S.C., Chiba, S., Suzuki, M., Su, E., Roberson, E.C., Pusapati, G.V., Setou, M., Rohatgi, R., Reiter, J.F., Ikegami, K., et al. (2017). Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168(1-2): 264-279 e215.
- Pugacheva, E. N., Jablonski, S. A., Hartman, T. R., Henske, E. P. and Golemis, E. A. (2007). HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129(7): 1351-1363.
- Saito, M., Otsu, W., Hsu, K. S., Chuang, J. Z., Yanagisawa, T., Shieh, V., Kaitsuka, T., Wei, F. Y., Tomizawa, K. and Sung, C. H. (2017). Tctex-1 controls ciliary resorption by regulating branched actin polymerization and endocytosis. EMBO Rep 18(8): 1460-1472.
- Shnitsar, I., Bashkurov, M., Masson, G. R., Ogunjimi, A. A., Mosessian, S., Cabeza, E. A., Hirsch, C. L., Trcka, D., Gish, G., Jiao, J., Wu, H., Winklbauer, R., Williams, R. L., Pelletier, L., Wrana, J. L. and Barrios-Rodiles, M. (2015). PTEN regulates cilia through Dishevelled. Nat Commun 6: 8388.
- Tomoshige, S., Kobayashi, Y., Hosoba, K., Hamamoto, A., Miyamoto, T., and Saito, Y. (2017). Cytoskeleton-related regulation of primary cilia shortening mediated by melanin-concentrating hormone receptor 1. Gen Comp Endocrinol 253(1): 44-52
- Tucker, R. W., Pardee, A. B. and Fujiwara, K. (1979). Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell 17(3): 527-535.
- Yeh, C., Li, A., Chuang, J. Z., Saito, M., Caceres, A. and Sung, C. H. (2013). IGF-1 activates a cilium-localized noncanonical Gbg signaling pathway that regulates cell-cycle progression. Dev Cell 26(4): 358-368.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Saito, M., Sakaji, K., Otsu, W. and Sung, C. (2018). Ciliary Assembly/Disassembly Assay in Non-transformed Cell Lines. Bio-protocol 8(6): e2773. DOI: 10.21769/BioProtoc.2773.
Category
Developmental Biology > Cell growth and fate > Proliferation
Developmental Biology > Cell signaling > Ligand
Cell Biology > Cell-based analysis > Organelle motility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









