- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Bacterial Attachment to Tissue Sections
Published: Vol 8, Iss 5, Mar 5, 2018 DOI: 10.21769/BioProtoc.2741 Views: 8761
Reviewed by: Andrea PuharSofiane El-Kirat-ChatelMigla Miskinyte

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2901 Views

A Novel Optimized Silver Nitrate Staining Method for Visualizing and Quantifying the Osteocyte Lacuno-Canalicular System (LCS)
Jinlian Wu [...] Libo Wang
Apr 20, 2025 1520 Views

An Ex Vivo Lung Histoculture Model for Studying Pulmonary Infection and Immune Response With SARS-CoV-2 as an Example of RNA Virus
Elena V. Maryukhnich [...] Elena J. Vasilieva
Dec 20, 2025 740 Views
Abstract
Here we describe a method to test bacterial adhesion to paraffin embedded tissue sections. This method allows examining binding of different bacterial strains, transfected with a fluorescent protein reporter plasmid to various tissues, to better understand different mechanisms such as colonization. This assay provides a more physiological context to bacterial binding, than would have been achieved using adhesion assays to cell lines. The sections can be imaged using fluorescent microscopy and adhesion of various bacterial strains can be quantified and tested, simultaneously.
Keywords: Host-pathogen interactionsBackground
Many types of bacteria, both commensal and pathogenic, express various adhesion molecules, allowing them binding to different surfaces of the host (Gur et al., 2015; Abed et al., 2016; Isaacson et al., 2017). This adhesion is crucial, as it is the first step of colonization and plays a role in both competition and survival, in different environments (Schilling et al., 2001). Many of these adhesins are lectins, binding sugar moieties on glycoproteins on various kinds of cells, such as epithelial cells and others (Abed et al., 2016; Isaacson et al., 2016). Over the years, many groups studying host-pathogen interactions used cell lines and tissue culture in order to try to understand bacterial adhesion to cells. Tissue sections give a more physiological context to the colonization study, as they provide organization and structures that are almost impossible to obtain using in vitro tissue culture. Furthermore, in immortalized or cancerous cells, the expression pattern of surface molecules, to which bacteria can bind, might be altered. In order to better understand physiological context of bacterial adherence, in both normal and pathological conditions, we chose to employ bacterial attachment to tissue sections.
Materials and Reagents
- Plastic 50 ml tubes for centrifugation (Greiner Bio One International, catalog number: 227270 )
- 1.5 ml tubes for transformation
- Petri dishes for bacteria (FL MEDICAL, catalog number: 29052 )
- Inoculation loop, 10 μl (Greiner Bio One International, catalog number: 731171 )
- Ice box with ice
- Slide jars for washing
- Superfrost Plus glass slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: J1800AMNT )
- Coverslip (Bar Naor, catalog number: BNBB024050A1 )
- Pipette tips (20-200 μl, 100-1,000 μl)
- Escherichia coli strain of interest (for example CFT073)
- Plasmids for fluorescent protein reporter expression (see references for examples)
- Calcium chloride (Sigma-Aldrich, catalog number: C5670 )
- Glycerol anhydrous (Avantor Performance Materials, J.T. Baker, catalog number: 2136 )
- Phospho-buffered saline (PBS 10x) (HyLabs, catalog number: BP-507/1Ld )
- Paraformaldehyde (PFA) (Bar Naor, catalog number: BN15711 )
- Xylene (Sigma-Aldrich, catalog number: 534056 )
- Ethanol (Sigma-Aldrich, catalog number: E7023 )
- ProLongTM Glass Antifade Mountant (Thermo Fisher Scientific, InvitrogenTM, catalog number: P36980 )
- Hoechst 33258 (Sigma-Aldrich, catalog number: 94403 )
- Dehydrated culture media, LB Broth (BD, DifcoTM, catalog number: 244620 )
- Agar purified for microbiology (Sigma-Aldrich, catalog number: 05038 )
- Erythromycin (Sigma-Aldrich, catalog number: E6376 )
- Ampicillin (Bio Basic, catalog number: AB0028 )
- Tris (Avantor Performance Materials, J.T. Baker, catalog number: 4109-1 )
- Sodium chloride (Avantor Performance Materials, J.T. Baker, catalog number: 3624-19 )
- Polyoxyethylene 20 sorbitan monolaurate (Tween 20) (Sigma-Aldrich, catalog number: 93774 )
- Bovine serum albumin (BSA) (VWR, Ameresco, catalog number: 97061-420 )
- Fetal bovine serum (FBS) (Biological Industries, catalog number: 04-0071A )
- Triton X-100 (Avantor Performance Materials, J.T. Baker, catalog number: X198-07 )
- LB medium (see Recipes)
- LB agar plates with antibiotics (see Recipes)
- TBSS solution (10x) (see Recipes)
- Blocking solution (see Recipes)
Equipment
- Pipettes
- Autoclave
- Spectrophotometer (600 nm wavelength)
- Shaker
- Micro centrifuge
- Incubator
- Thermoblock
- Chemical hood
- Fluorescence microscope (TL-Nikon)
Software
- ImagePro Analyzer 7.0 software
- Software for statistical analysis (GraphPad Prism software version 6.0 or later, for example)
Procedure
The procedure outline is described in Figure 1.
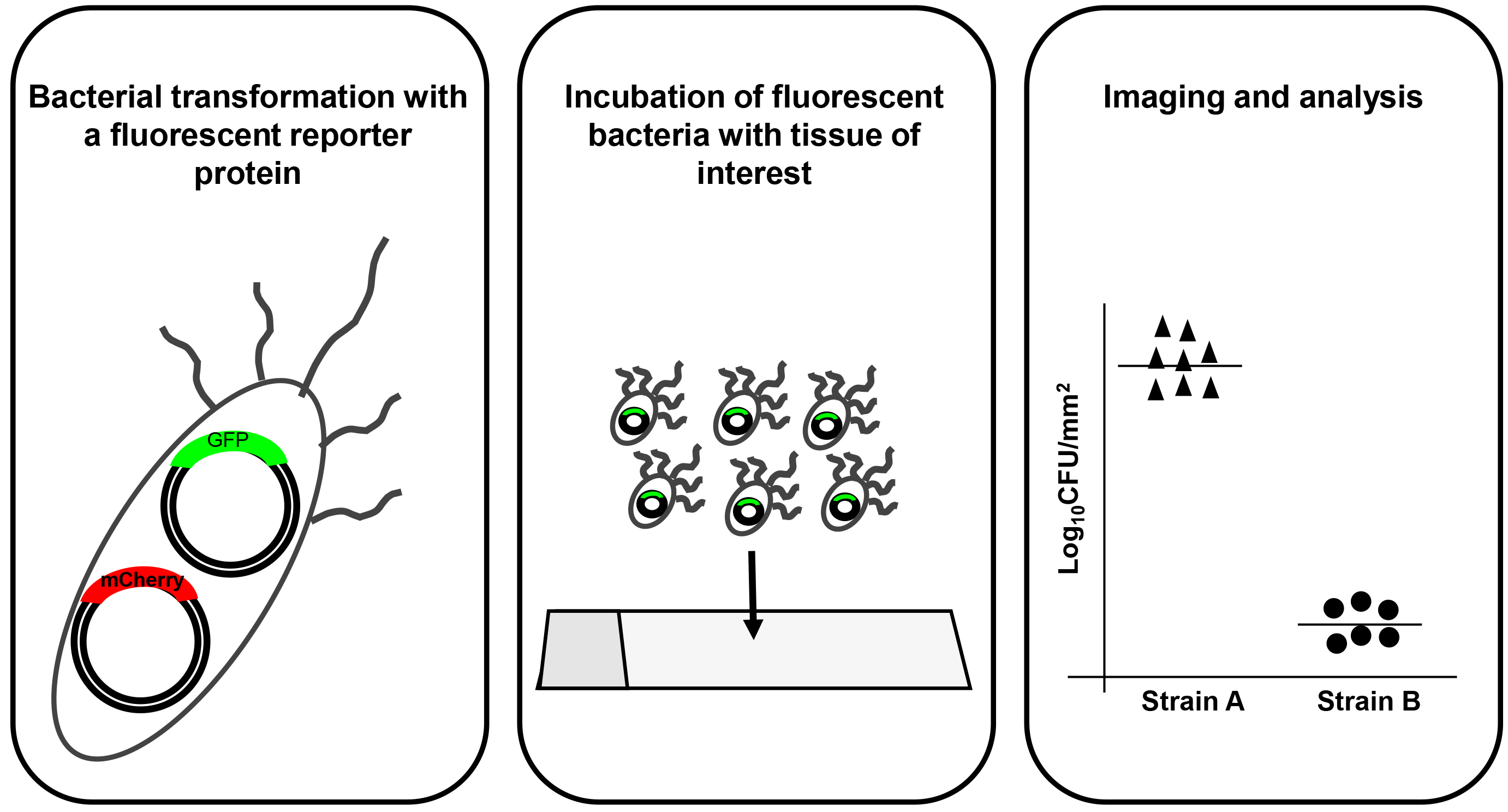
Figure 1. Protocol outline. General outline of the protocol describing the three main stages of the assay. The left panel shows preparation of fluorescent protein expressing E. coli. The middle panel outlines the tissue adhesion test and the right panel shows an output of analysis of data acquired during imaging.
- Preparation of competent bacteria
- Grow E. coli strain of interest in 5 ml of sterile LB medium (see Recipes) in a 50 ml tube, overnight (12-18 h) at 220 rpm shaking at 37 °C.
- Inoculate 500 μl of the overnight starter culture into 50 ml of preheated LB (37 °C) and grow for two hours, at 220 rpm shaking at 37 °C until OD600 of 0.3 to 0.4.
- Centrifuge at 4 °C, 3,220 x g for 10 min.
- Discard supernatant, keep pellet on ice for 10 min.
- Suspend pellet in 20 ml of 0.1 M cold CaCl2.
- Leave for 25 min on ice.
- Centrifuge again as indicated in Step A3, discard supernatant and suspend pellet in 2 ml of 0.1 M CaCl2 + 15% glycerol.
Note: Glycerol should be autoclaved and the CaCl2 solution should be filtered prior to use.
- Incubate on ice for 90 min.
- Grow E. coli strain of interest in 5 ml of sterile LB medium (see Recipes) in a 50 ml tube, overnight (12-18 h) at 220 rpm shaking at 37 °C.
- Bacterial transformation
- Take 100 μl of competent bacteria into a 1.5 ml tube and add 30 ng of the plasmid of choice encoding for–either GFP (Hansen et al., 2001) or mCherry (Sason et al., 2009).
- Incubate for 20 min on ice.
- Transfer tubes to a thermoblock heated to 42 °C for 90 sec.
- Move tubes to ice for 5 min.
- Add 1 ml sterile LB and shake for 1 h at 37 °C, 220 rpm.
- Centrifuge at 4,830 x g for 5 min.
- Resuspend pellet in 150 μl of fresh LB and seed on an LB agar plate supplemented with appropriate antibiotic for selection, according to resistance encoded on the plasmid of choice (here ampicillin and erythromycin, see Recipes).
- Incubate plate overnight at 37 °C.
- The next day–pick a single colony, grow in 5 ml LB (supplemented with appropriate antibiotics, see Recipes section) overnight (12-18 h) at 220 rpm shaking at 37 °C.
Note: In order to avoid loss of fluorescent signal, it is strongly recommended that bacteria expressing fluorescent proteins should be protected from light at all times.
- Bring bacterial culture to OD600 of 1 (dilute in sterile 1x PBS).
- Take 100 μl of competent bacteria into a 1.5 ml tube and add 30 ng of the plasmid of choice encoding for–either GFP (Hansen et al., 2001) or mCherry (Sason et al., 2009).
- Tissue binding assay
This assay uses 4 μm thick paraffin embedded section of tissues fixated in PFA, mounted on glass slides (see Materials and Reagents).- Fill three staining jars with these three solutions and perform deparaffinization as described (Figure 2A):
- Xylene–5 min, 5 min, 2 min.
- Ethanol 100%–5 min, 5 min, 2 min.
- Ethanol 96%–3 min, 2 min, 2 min.
- Xylene–5 min, 5 min, 2 min.
- Cover sections with blocking solution (see Recipes) and incubate at room temperature for 6 h.
- Suspend 50 μl of bacteria at OD600 = 1 in 950 μl blocking solution, after discarding blocking solution, lay the bacterial suspension gently on slide.
- Incubate overnight at 4 °C in a wet chamber (line chamber with wet tissues), protected from light.
- Prepare 2 staining jars filled with 1x PBS and another staining jar containing PBS with 0.05% Tween 20 (PBST, Figure 2B).
- Wash twice with PBS for 5 min per wash. Transfer the slides to PBST and wash for 10 min (Figure 2B).
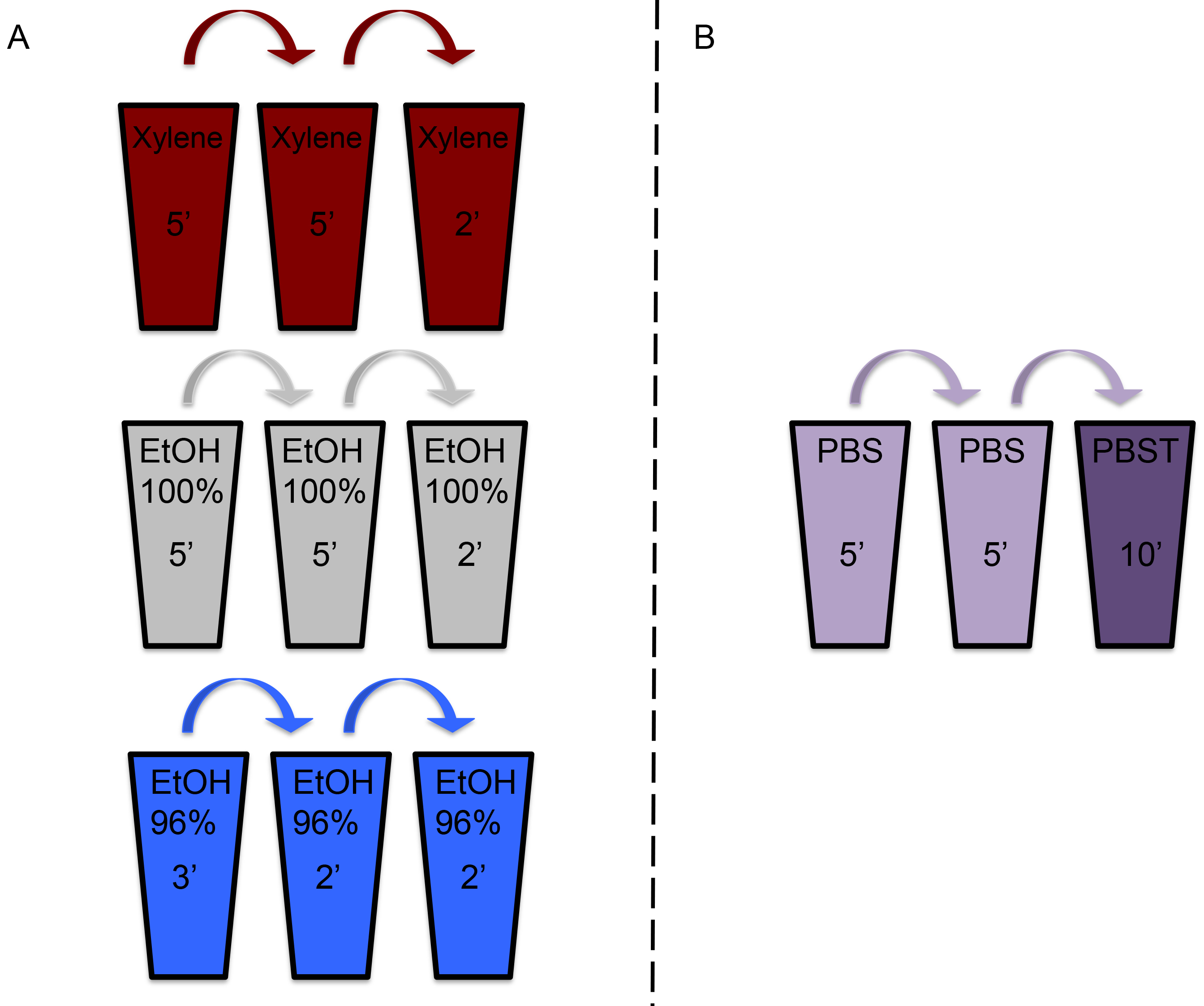
Figure 2. Washing/deparaffinization illustration. Prepare three washing jars filled with the solution indicated on the jar in the illustration and keep the slides in the jar for the indicated amount of time. Figure 2A illustrates the first round of slide deparaffinization in preparation for the binding assay. Figure 2B illustrates washing after overnight incubation to wash off unbound bacteria, dark purple jar contains PBS supplemented with tween (PBST).
- Dilute Hoechst 33258 1:5,000 in 1x PBS at approximately 200 μl per slide, apply and incubate for 20 min at room temperature. Protect from light.
- Apply mounting medium to slide and cover with a coverslip.
- Imagining can be done under a fluorescence microscope using a 60x magnification.
Note: Scan at least four fields per slide.
- Fill three staining jars with these three solutions and perform deparaffinization as described (Figure 2A):
Data analysis
- Images obtained from the fluorescence microscope are converted to 8 bit images by fluorescence microscopy image analysis software (see Figure 3, for example).
- Fluorescent bacteria should be quantified (for each field) by two different experimenters for a total tissue area of 1,600 µm2.
- Convert field to mm2. Each fluorescent bacterium counted represents a colony forming unit (CFU), data are represented as Log10CFU/mm2.
Example: For convenience purposes, this example will refer to a field of 100 µm2.
(CFU in 100 µm2) = (CFU in 0.1 mm2)
10x (CFU in 100 µm2) = CFU in 1 mm2
If the counted CFU in 100 µm2 is 1,000, the CFU in mm2 will be 10,000 (or, 104) and therefore the log10CFU/mm2 is 4.
- Each spot of a single bacterium is referred to as a CFU.
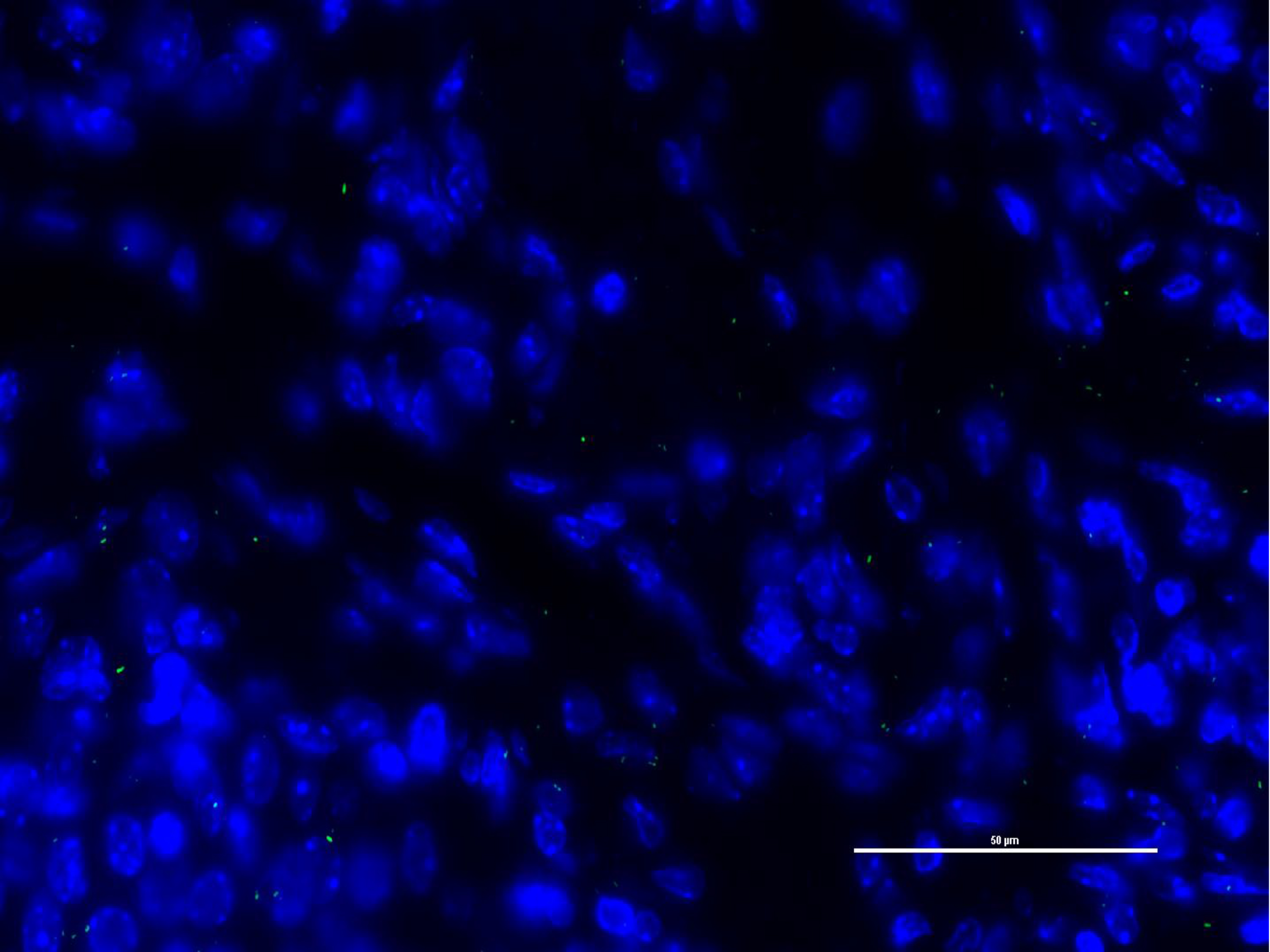
Figure 3. Fluorescent microscope image of GFP expressing uropathogenic E. coli (UPEC) adhesion paraffin embedded bladder tissue section. Scale bar = 50 µm.
Recipes
- LB medium
1 L of double distilled water (DDW)
20 g LB dehydrated culture media
Mix until dissolved
Autoclave at 121 °C for 30 min and aliquot
- LB agar plates with antibiotics
1 L of DDW
20 g LB dehydrated culture media
15 g of purified agar
Mix and autoclave. Agar will dissolve during autoclave heating
Let cool until LB agar can be handled, before it gets solidified
Add antibiotics (erythromycin at 6 mg/ml and ampicillin at 1 mg/ml) and pour plates
- TBSS solution (10x)
500 ml 0.5 M Tris (pH 7.4)
800 ml 2 M NaCl
2 ml Tween 20
Add DDW up to 2 L
Mix well
- Blocking solution
100 ml 1x TBSS
15 g BSA
15 ml FBS
5.75 ml 5% Triton X-100
Acknowledgments
This study was supported by the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP/2007-2013) ERC grant 320473-BacNK. Further support came from the I-CORE Program of the Planning and Budgeting Committee and the Israel Science Foundation and by the I-Core on Chromatin and RNA in Gene Regulation, the GIF Foundation, the Lewis Family Foundation, an ICRF professorship grant, a Helmholtz Israel grant, a Kamin grant, and the Rosetrees Trust (all to O.M.). O.M is a Crown professor of Molecular Immunology. The authors declare no conflict of interests.
References
- Abed, J., Emgard, J. E., Zamir, G., Faroja, M., Almogy, G., Grenov, A., Sol, A., Naor, R., Pikarsky, E., Atlan, K. A., Mellul, A., Chaushu, S., Manson, A. L., Earl, A. M., Ou, N., Brennan, C. A., Garrett, W. S. and Bachrach, G. (2016). Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host Microbe 20(2): 215-225.
- Gur, C., Ibrahim, Y., Isaacson, B., Yamin, R., Abed, J., Gaml iel, M., Enk, J., Bar-On, Y., Stanietsky-Kaynan, N., Coppenhagen-Glazer, S., Shussman, N., Almogy, G., Cuapio, A., Hofer, E., Mevorach, D., Tabib, A., Ortenberg, R., Markel, G., Miklic, K., Jonjic, S., Brennan, C. A., Garrett, W. S., Bachrach, G. and Mandelboim, O. (2015). Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42(2): 344-355.
- Hansen, M. C., Palmer, R. J., Jr., Udsen, C., White, D. C. and Molin, S. (2001). Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147(Pt 5): 1383-1391.
- Isaacson, B., Hadad, T., Glasner, A., Gur, C., Granot, Z., Bachrach, G. and Mandelboim, O. (2017). Stromal cell-derived factor 1 mediates immune cell attraction upon urinary tract infection. Cell Rep 20(1): 40-47.
- Sason, H., Milgrom, M., Weiss, A. M., Melamed-Book, N., Balla, T., Grinstein, S., Backert, S., Rosenshine, I. and Aroeti, B. (2009). Enteropathogenic Escherichia coli subverts phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate upon epithelial cell infection. Mol Biol Cell 20(1): 544-555.
- Schilling, J. D., Mulvey, M. A. and Hultgren, S. J. (2001). Structure and function of Escherichia coli type 1 pili: new insight into the pathogenesis of urinary tract infections. J Infect Dis 183 (Suppl 1): S36-S40.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Isaacson, B., Hadad, T., Bachrach, G. and Mandelboim, O. (2018). Quantification of Bacterial Attachment to Tissue Sections. Bio-protocol 8(5): e2741. DOI: 10.21769/BioProtoc.2741.
Category
Immunology > Host defense > Murine
Microbiology > Microbe-host interactions > Ex vivo model
Cell Biology > Tissue analysis > Tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link