- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Characterization of Amyloid Fibril Networks by Atomic Force Microscopy
Published: Vol 8, Iss 4, Feb 20, 2018 DOI: 10.21769/BioProtoc.2732 Views: 9012
Reviewed by: Vivien Jane Coulson-ThomasVijaykrishna RaghunathanAnca Flavia Savulescu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Protein Vesicle Tracking in HeLa Cells
Natalia Pacheco-Fernandez [...] Julia von Blume
Mar 5, 2021 9242 Views

Patterned Substrate of Mobile and Immobile Ligands to Probe EphA2 Receptor Clustering
Zhongwen Chen [...] Jay T. Groves
Jun 5, 2022 3043 Views

In vitro Reconstitution of Phase-separated p62 Bodies on the Arp2/3-derived Actin Network
Tong Liu [...] Na Mi
Apr 20, 2023 2236 Views
Abstract
Dense networks of amyloid nanofibrils fabricated from common globular proteins adsorbed to solid supports can improve cell adhesion, spreading and differentiation compared to traditional flat, stiff 2D cell culture substrates like Tissue Culture Polystyrene (TCPS). This is due to the fibrous, nanotopographic nature of the amyloid fibril networks and the fact that they closely mimic the mechanical properties and architecture of the extracellular matrix (ECM). However, precise cell responses are strongly dependent on the nanostructure of the network at the cell culture interface, thus accurate characterization of the immobilized network is important. Due to its exquisite lateral resolution and simple sample preparation techniques, Atomic Force Microscopy (AFM) is an ideal technique to characterize the fibril network morphology. Thus, here we describe a detailed protocol, for the characterization of amyloid fibril networks by tapping mode AFM.
Keywords: Amyloid nanofibrilsBackground
Networks of non-toxic amyloid fibrils assembled from common globular proteins (Jung et al., 2008; Lara et al., 2011) adsorbed to solid supports have applications in a wide variety of fields (Dharmadana et al., 2017; Wei et al., 2017). Particularly interesting is their applications in eukaryotic cell culture and biomaterials in general (Reynolds et al., 2013, 2014 and 2015; Gilbert et al., 2017b). This is largely due to the fact that networks of amyloid fibrils have morphologies and mechanical properties that closely resemble the local microenvironment of many cell types (the ECM). Such amyloid fibril networks have the added attraction that they are simple to fabricate, inexpensive and possess well-defined chemistries that can be easily reproduced.
As expected, the response of cells grown on these amyloid fibril networks is highly dependent on the nanoscale properties of the fibril network itself. For instance, small changes in fibril diameter, nanoscale roughness, surface coverage and fibril morphology have been shown to affect cell attachment and spreading (Reynolds et al., 2014 and 2015). Thus, it is important to accurately characterize the nanotopography and surface roughness of the immobilized networks before using them for cell culture applications. AFM is a powerful technique to perform this analysis as it requires little sample preparation, possesses nanoscale lateral resolution and sub-nanometer vertical resolution (Reynolds et al., 2014 and 2015; Gilbert et al., 2017a and 2017b; Reynolds et al., 2017). Additionally, parameters such as nanoscale roughness can be extracted by post imaging analysis. In this protocol, we will describe the process of imaging a dense network of amyloid fibrils (fabricated from the protein Hen Egg White Lysozyme) on solid (mica) substrates by AFM. We will also describe the most common steps of post-processing analysis, namely flattening (removal of sample tilt and bowing artefacts) and roughness analysis.
Materials and Reagents
- AFM Metal Specimen discs Diameter 15 mm (ProSciTech, catalog number: GA530-15 )
- Muscovite Mica disks, grade V-1 diameter 12.5 mm (ProSciTech, catalog number: G51-12 )
- STKYDOT adhesive pads (Bruker Nano, catalog number: STKYDOT )
Equipment
- Cole-Parmer Precision Tweezer Set, Stainless Steel (Cole-Parmer Instrument, catalog number: 07387-16 )
- Multimode 8 Atomic Force Microscope (AFM) with Nanoscope V controller (Bruker Nano, model: Multimode 8 )
- Tapping Mode AFM tips (Approx. Resonant Frequency = 300 kHz, force constant 40 N/m) (Bruker Nano, model: RTESPA-300 )
Software
- Nanoscope Analysis Software (Bruker Version 1.7)
Procedure
Note: All procedures here are described for a Bruker Multimode AFM, for different models and brands of AFM the protocol will need to be adapted according to the manufactures instructions.
- Loading the sample and setting up the AFM
Modern AFM instruments have a multitude of different imaging modes to choose from including contact mode, non-contact mode and tapping mode. For delicate soft materials or biomaterials such as this, it is important to minimize the force exerted on the substrate by the AFM tip to prevent the surface being damaged. Tapping mode AFM (TM-AFM) achieves this by intermediately tapping the substrate, and not dragging the hard, sharp tip of the AFM cantilever across the substrate as in contact mode AFM. For this reason, all imaging on these amyloid fibril networks should be performed in TM-AFM.- Amyloid fibril networks are fabricated by exposing globular proteins (typically β-lactoglobulin or hen egg white lysozyme) to high temperatures and low pH which causes the proteins to be hydrolyzed into peptide fragments.Over time the fragments self-assemble into amyloid fibrils (see Charnley et al. [2018] for a detailed protocol on fabricating these networks). The freshly prepared amyloid fibril networks can be deposited on mica via a simple drop-casting protocol (see Charnley et al. [2018]) and can now be attached to the magnetic AFM stub using one of the double-sided sticky dots. Carefully mount the sample onto the AFM (Figure 1a) ensuring not to damage the mirror in the top left of the sample loading area.
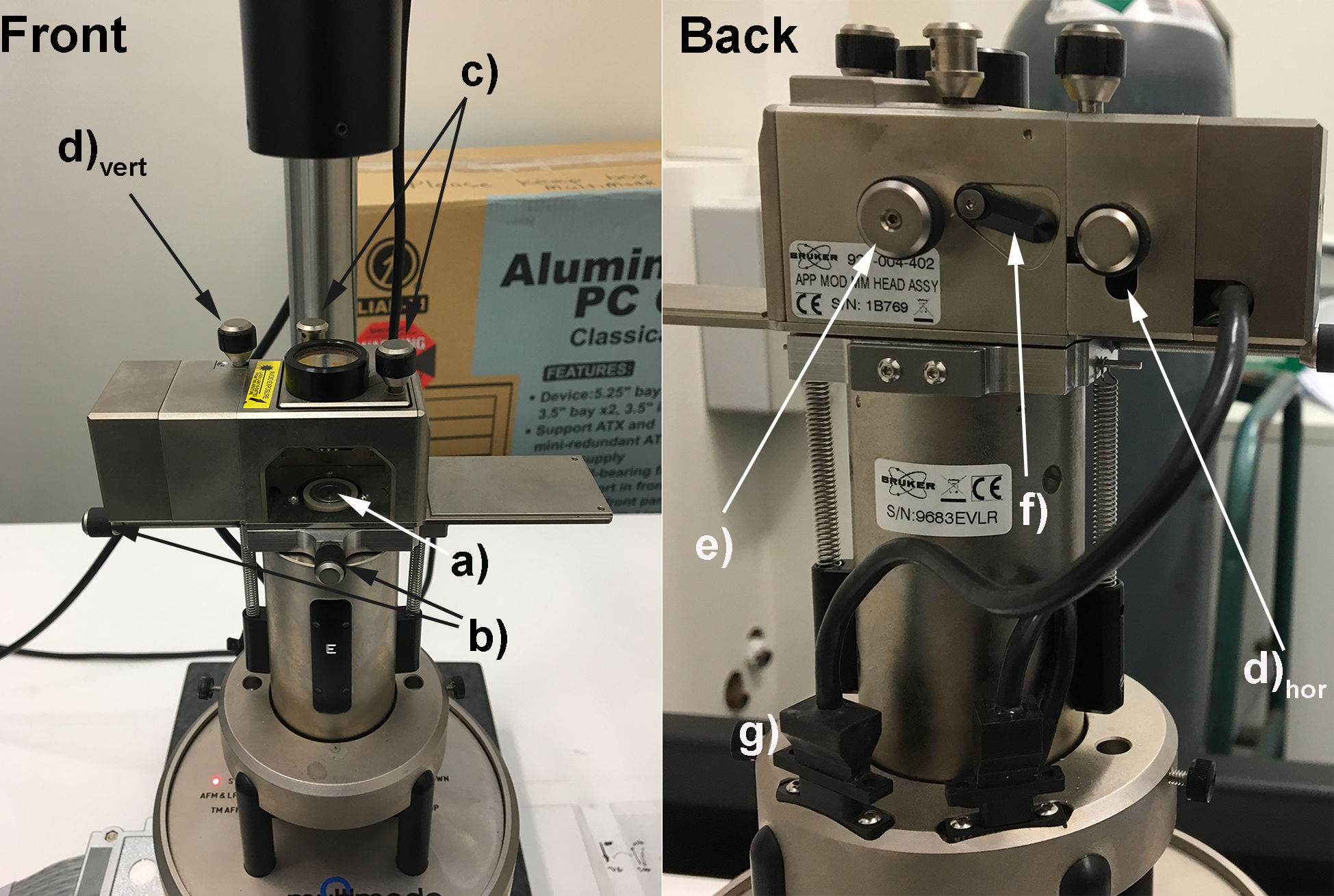
Figure 1. Bruker Multimode 8 AFM with important controls labelled. a) Magnetic sample holder (for samples attached to magnetic AFM stubs; b) Knobs to manipulate x-y stage; c) Knobs to manipulate laser position; d)vert Knob to manipulate the vertical displacement of the photodiode; d)hor Knob to manipulate the horizontal displacement of the photodiode; e) Knob to secure and unsecure the cantilever holder; f) Lever to adjust angle of mirror directing laser onto the photodiode; g) Cable and connector for the laser source (ensure it is plugged in). - Mount the AFM cantilever into its holder, ensuring that the tip is not damaged (Video 1), and that the base of the cantilever sits flush with the back of the cantilever holder (Figure 2a). Now load the AFM cantilever holder into the AFM, ensuring that the sample is not too high, which would cause the cantilever to crash into the sample. Ensure that the AFM is in AFM mode (not STM or TM-AFM).
Video 1. Loading AFM cantilever into the cantilever holder - Focus the optical microscope on the AFM to the cantilever and ensure that it is mounted straight and not at an angle (Figures 2b and 2c). Refocus the camera on the surface of the mica sheet.
Note: Take care that you are focussed on the uppermost surface, to prevent crashing the cantilever into the substrate. - Move the sample upwards (or AFM tip downwards depending on the AFM model) so that it is close to the mica substrate, the distance from the substrate can be judged by looking at the separation between the AFM tip and its reflection (Figure 2d).
- Focus the AFM laser spot on the cantilever (Figure 1c), maximizing the obtained Sum (it should be around 7 for an RTESPA300 tip), but also ensuring that the laser spot is close to the tip of the cantilever and not significantly over one edge (Figure 2e).
Note: To maximize the Sum both the laser position (Figure 1c) and the angle of the mirror (Figure 1f) that focusses the laser onto the 4 quadrant photodiode within the AFM may need to be adjusted. - After correctly positioning the laser spot onto the AFM cantilever, the 4 quadrant photodiode should be adjusted so that both the vertical and horizontal deflection reads as zero (Figure 1dvert and 1dhor). Once correctly adjusted the AFM should be switched to TM-AFM mode (using the switch on the base of the instrument) and the horizontal deflection reset to zero if required.
Note: Upon switching to TM-AFM the vertical deflection will have disappeared and have been replaced with the RMS value.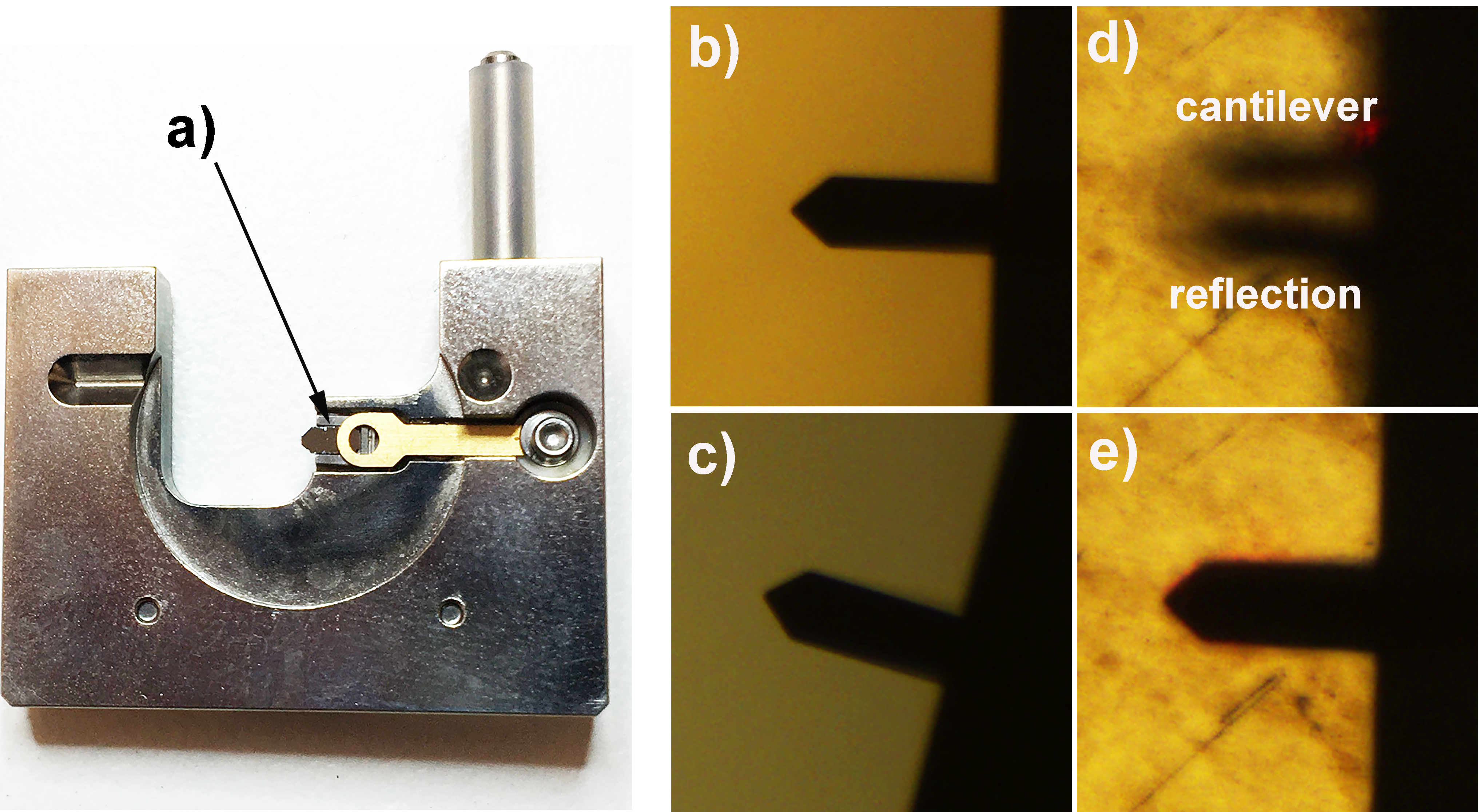
Figure 2. Cantilever and laser alignment. a) Correctly mounted RTESPA300 cantilever in a standard Bruker multimode cantilever holder; b) Optical microscopy image of correctly aligned cantilever; c) Optical microscopy image of poorly aligned cantilever; d) Optical microscope image of cantilever approaching the surface, tip-sample separation can be monitored by observing the gap between the image of the cantilever and its reflection; e) When the cantilever is sufficiently close to the surface the image of the cantilever and its reflection cannot be distinguished from each other, at this point the rest of the approach should be performed by the software (using the engage command). - Now the cantilever is correctly mounted and the laser aligned, all that remains before imaging is to select the resonant frequency of the cantilever. This can be done in the setup menu of the AFM software (within the soft tapping mode experimental pre-set). As this imaging is being performed in air, the autotune software (Figure 3) works well however, the user should ensure that autotune is set up to scan across the correct frequency range (200-400 kHz works well for the RTESPA300 cantilevers) and that the target amplitude is sufficient (500 mV is a good place to start) (see Figure 3).
Note: When tuning the cantilever to find the precise resonant frequency ensure that the cantilever is sufficiently far away from the surface so that the resonant frequency is not influenced by sample-tip interactions.
Upon clicking execute in the autotune menu, the software should correctly identify the resonant frequency of the cantilever (likely to be somewhere between 270-330 kHz for RTESPA300 cantilevers) and fix the amplitude to the chosen target amplitude (see Figure 3). If the software cannot identify a resonant frequency or it looks suspicious (i.e., > ± 50 kHz from the quoted resonant frequency, and/or not obviously the most intense peak) then Steps A2-A5 may need to be repeated or the cantilever may be damaged.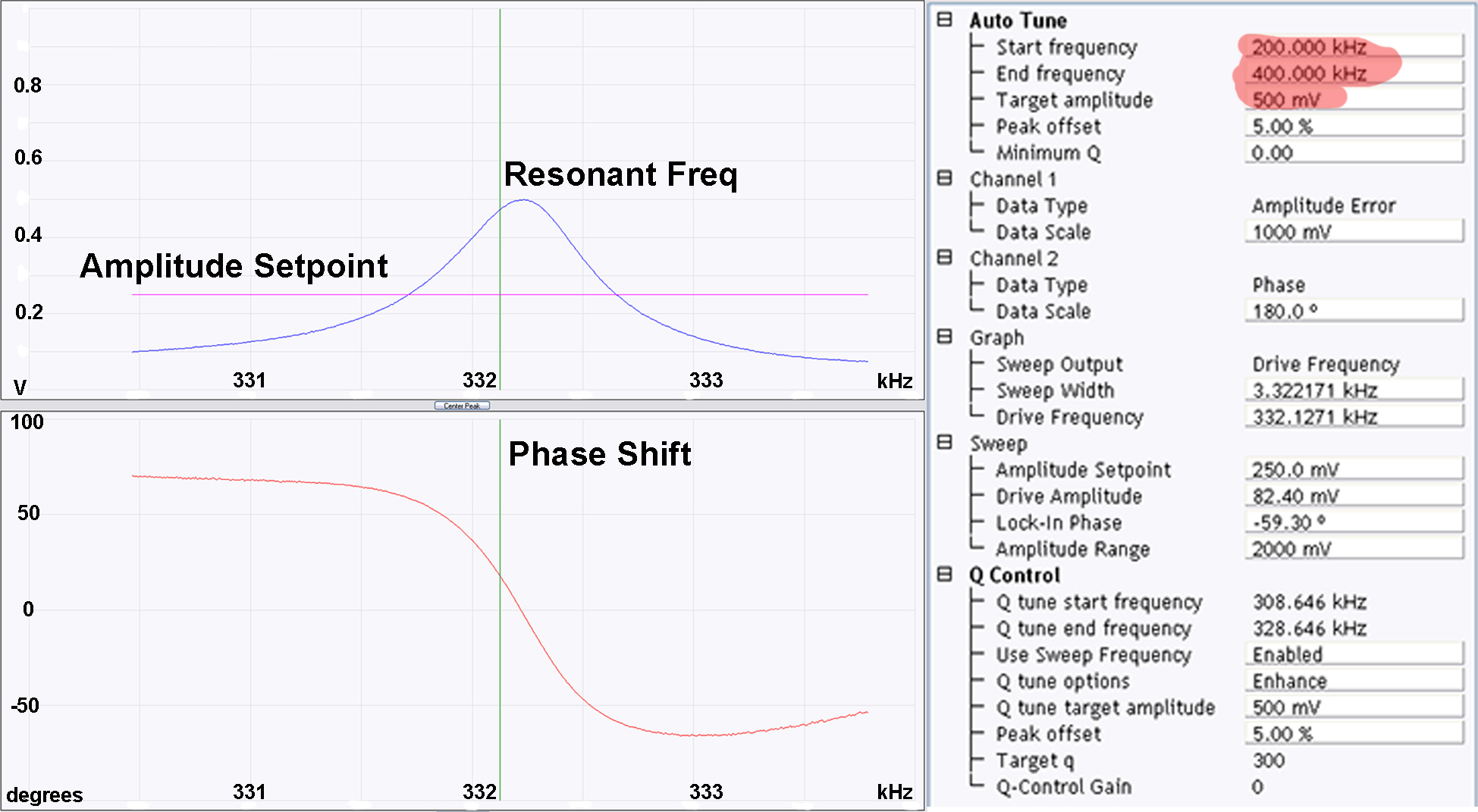
Figure 3. Cantilever tuning. The resonant frequency and phase shift should look approximately like the examples given above with one major peak around (± 50 kHz) the quoted resonant frequency of the cantilever (in this case 300 kHz). Particular attention should be given to the highlighted parameters, the start and end frequency should include the expected resonant frequency (300 kHz) and the desired target amplitude should be selected (for amyloid fibrils 500-1,000 mV works well).
- Amyloid fibril networks are fabricated by exposing globular proteins (typically β-lactoglobulin or hen egg white lysozyme) to high temperatures and low pH which causes the proteins to be hydrolyzed into peptide fragments.Over time the fragments self-assemble into amyloid fibrils (see Charnley et al. [2018] for a detailed protocol on fabricating these networks). The freshly prepared amyloid fibril networks can be deposited on mica via a simple drop-casting protocol (see Charnley et al. [2018]) and can now be attached to the magnetic AFM stub using one of the double-sided sticky dots. Carefully mount the sample onto the AFM (Figure 1a) ensuring not to damage the mirror in the top left of the sample loading area.
- Imaging the amyloid fibril networks
Maximum available resolution in the AFM will depend on a large variety of factors (tip quality, instrument noise, external noise, the piezo scanner used etc.). This protocol described uses a relatively high resolution ‘e’ scanner, which sacrifices available scan size for increased lateral resolution, thus maximum scan sizes are around 10 μm. Other piezo scanners and other models of AFM will allow for bigger scan sizes (up to hundreds of microns in some cases).- Ensure that the AFM cantilever is close to the substrate interface (Figure 2e), select a scan rate (~1 Hz), scan size (1-10 μm) and an image resolution (512 x 512 pixels for publication quality images) and engage the cantilever from the scan menu in the software. The sample will now automatically be moved (initially via a stepper motor, and later by a piezo motor), until the cantilever and substrate come into contact. Occasionally a false engage occurs when the cantilever fails to come into contact with the surface, if this happens multiple times then Steps A2-A5 above may need to be repeated or the tip may be damaged.
- The most important parameter to be optimised once the cantilever in contact with the substrate is the amplitude setpoint. The amplitude setpoint approximates to the force exerted by the tip on the sample (lower voltage = higher force exerted), and is automatically set to an initial value of 50% of the target amplitude. Once scanning has commenced the amplitude setpoint can be adjusted so that the tip closely tracks overall features on the surface. This can be assessed by looking at the trace and retrace line scans in the topography channel (Figure 4b). These should be exactly overlapping, if they are not then the amplitude setpoint should be reduced. An optimum amplitude setpoint is achieved when the AFM is just tracking the surface (in trace and retrace directions), but the force exerted on the sample is minimized (Figure 4). At this stage, if required, the drive amplitude and drive frequency can also be optimized to further improve the quality of the image.
- Finally adjust the integral (IG) and proportional gain (PG) so that the noise in all of the channels is minimised. The amplitude error channel (channel 2 by default) most clearly shows noise therefore this channel should be used to fine tune the IG and PG. Typically as the gains are increased a reduction in noise is observed until an upper limit is reached, beyond which additional electrical noise is introduced into the image. As a rule of thumb, the PG should always be 2-3 greater than the IG (Figure 4c). Once satisfied with the image quality, restart the scan, set the file name and directory and turn on capture to record the image. Examples of both good and bad scanning parameters are shown in Figure 4.
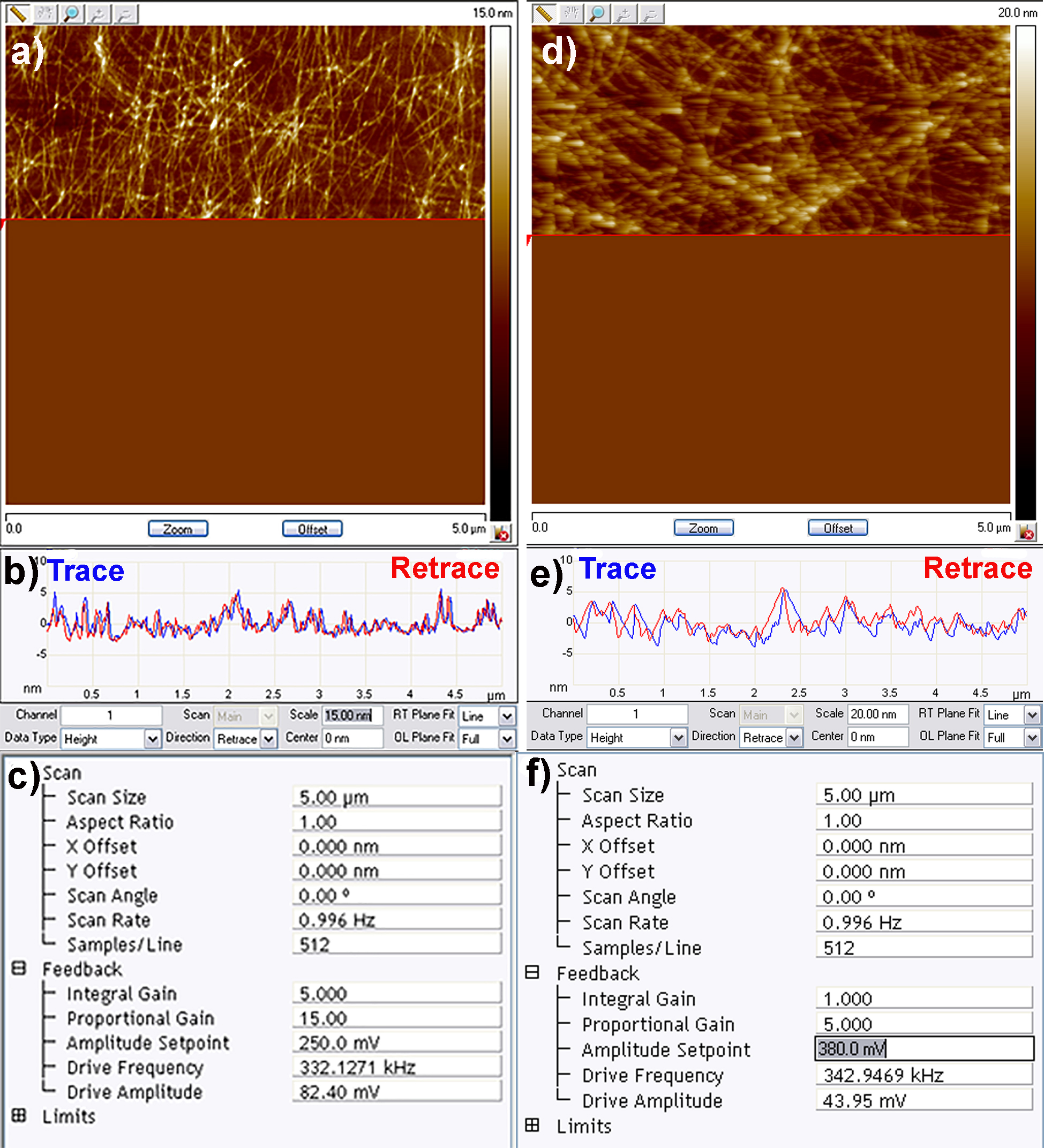
Figure 4. Good (left) and bad (right) quality AFM imaging of amyloid fibril networks. a) Example of amyloid fibril network image being recorded accurately; b) Trace and retrace scan lines overlapping confirming quality of the image; c) Typical scan settings used to record the above image; d) Poor imaging parameters resulting in the AFM cantilever not closely tracking the surface hence the blurry images; e) Trace and retrace scan lines not well overlapping due to high amplitude setpoint; f) Example of non-optimised scan settings.
- Ensure that the AFM cantilever is close to the substrate interface (Figure 2e), select a scan rate (~1 Hz), scan size (1-10 μm) and an image resolution (512 x 512 pixels for publication quality images) and engage the cantilever from the scan menu in the software. The sample will now automatically be moved (initially via a stepper motor, and later by a piezo motor), until the cantilever and substrate come into contact. Occasionally a false engage occurs when the cantilever fails to come into contact with the surface, if this happens multiple times then Steps A2-A5 above may need to be repeated or the tip may be damaged.
Data analysis
Basic image manipulation and roughness analysis
Here we will describe in detail the most basic post image processing used for the majority of AFM images. Note all processing and analysis protocols were performed in the Nanoscope Analysis software (Bruker v1.7), protocols may vary for other AFM analysis software.
- The first stage of image processing in all topographic AFM images is to correct for deviations from planarity (tilt due to samples not being mounted in the AFM perfectly horizontally). This is performed through the flatten tool in the Nanoscope Software (Figure 5). For images that scan a small percentage of the total possible distance of the piezo scanner, a first order flatten that simply corrects for tilt is sufficient. In the authors experience first order plane fits are appropriate up to approximately < 40% of the total available scanning distance, in the case of the multimode ‘e’ scanner this is roughly equal to 4-5 μm. For larger scans 2nd or 3rd order flatten algorithms may be required to remove both sample tilt and bowing artifacts that occur when the piezo travels over the majority of its working distance.
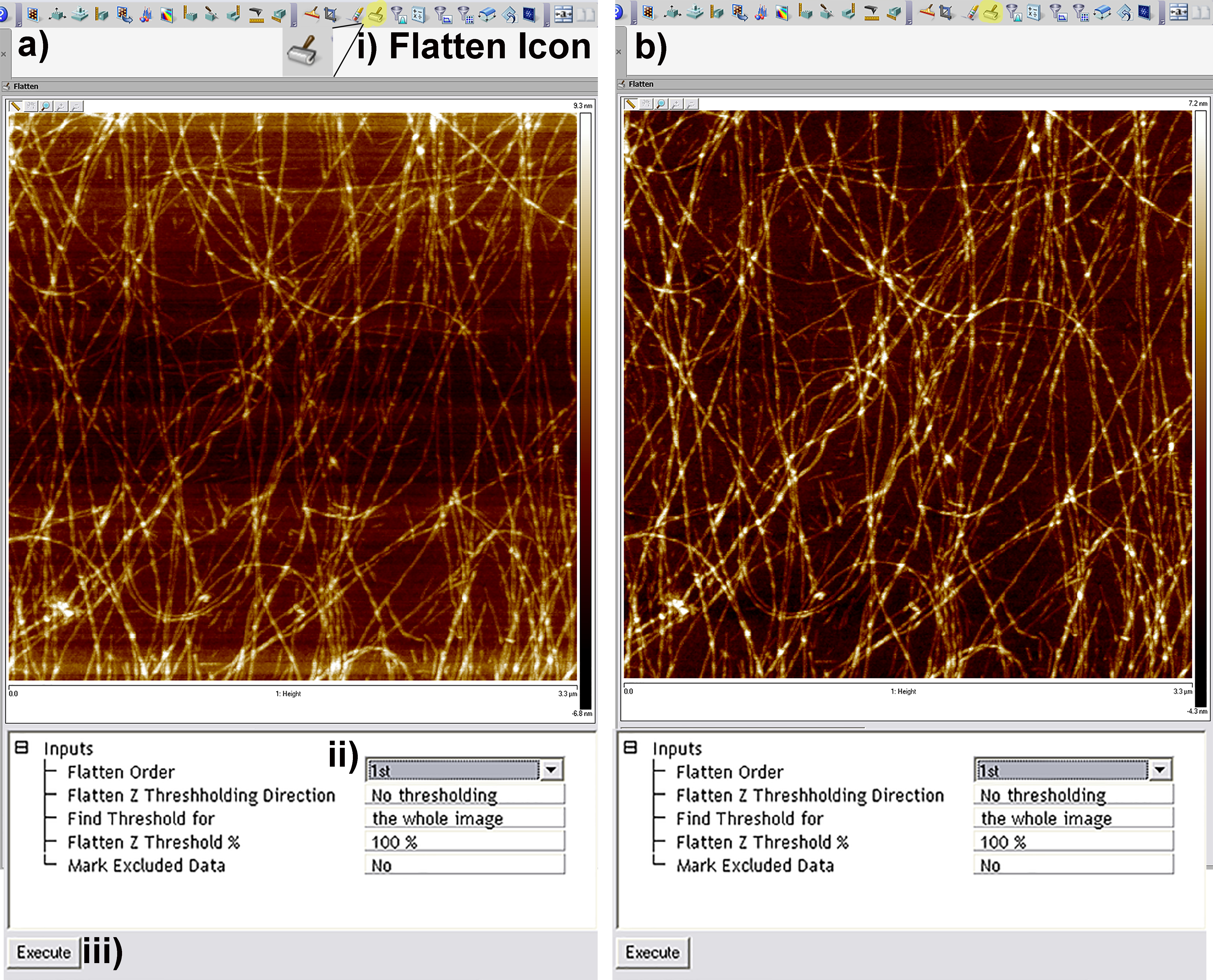
Figure 5. Screen Shots from the flattening tool in the Nanoscope Analysis Software. a) Pre-flattened image with multiple artifacts due to tilt in sample. b) 1st order flattened image removing tilt only. For larger scans 2nd or 3rd order flattening may be required to remove bowing effects caused by the piezo traveling a large percentage of its total possible distance. - In order to calculate both arithmetical and root mean square roughness (termed Ra and Rq respectively in the Nanoscope Analysis software), the roughness tool can be used by clicking on the roughness icon in the software (Figure 6i). The table that is generated as a result displays the Ra and Rq values for the entire image (Figure 6ii) and also for a specific region of interest (Figure 6iii) if selected by drawing a box on the image.
- There are many additional AFM analysis tools that can be used to pull out a wealth of information on amyloid fibril networks such as those described above, however it is beyond the scope of this protocol to describe them here. For a detailed description of all the tools available in Nanoscope Analysis see the help files associated with the software. In addition, for fibrillar materials, the authors recommend the free software FiberApp (Usov and Mezzenga, 2015) for more complex statistical analysis methods.
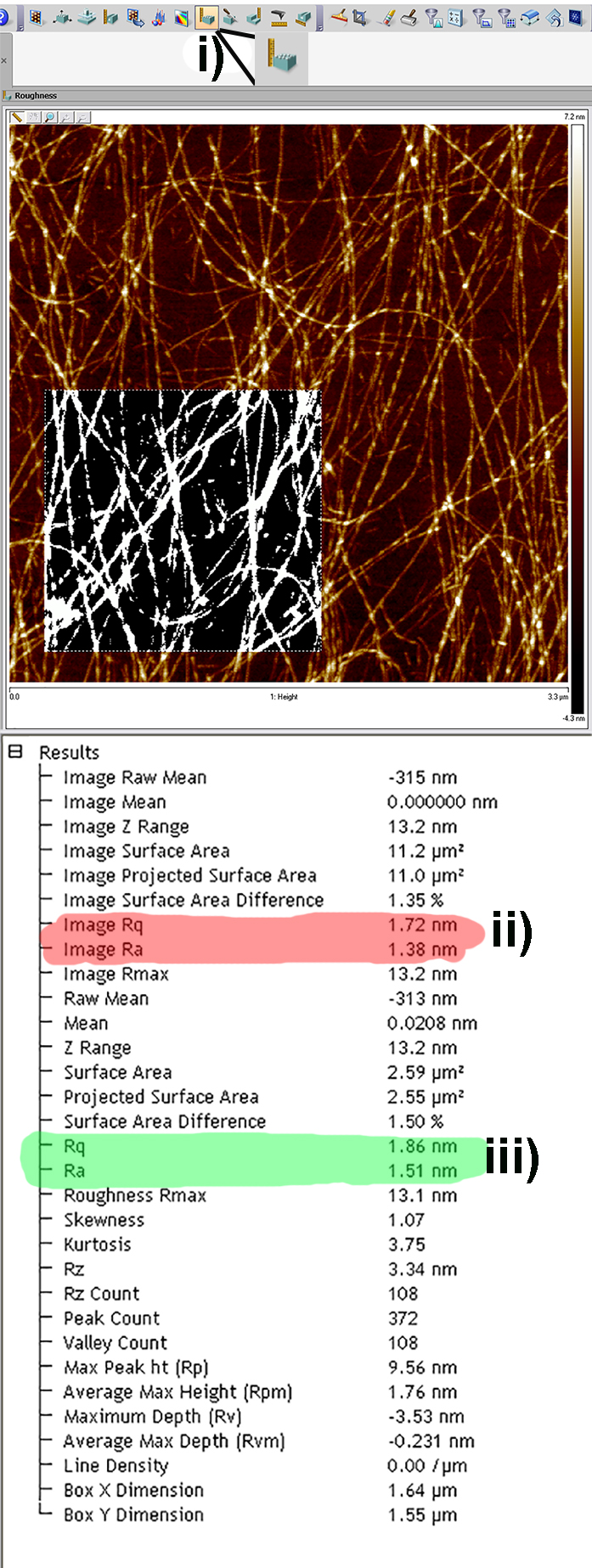
Figure 6. Screen Shots from the Roughness Analysis tool in the Nanoscope Analysis software. i) The roughness analysis icon; ii) highlighted in red are the Rq and Ra values for the entire of the image; iii) Highlighted in green are the Rq and Ra values for the selected region of the image.
Notes
The quality of the images obtained is highly dependent on the quality and cleanliness of the AFM cantilevers used, for results of publication quality we always recommend using pristine cantilevers and not cantilevers that have previously been used for imaging. Additionally, the quality of cantilevers from different manufacturers can vary massively, the authors note that often buying cantilevers from cheaper manufactures turns out to be economically unsound, as the quality is reduced to the point where many of the cantilevers are not usable.
Recipes
All the recipes for preparing the amyloid nanofibril networks imaged here are detailed in Charnley et al. (2018).
Acknowledgments
This work was performed in part at the ANFF-Vic node of the Australian National Fabrication Facility, a company established under the National Collaborative Research Infrastructure Strategy to provide nano-and micro-fabrication facilities for Australia’s researchers. JG acknowledges the Australian Government Department of Education and Training for an Endeavour Scholarship and the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1333468. NPR and JG acknowledge the ARC Training Centre for Biodevices at Swinburne University of Technology (IC140100023) for funding (NPR) and for hosting for the duration of his scholarship (JG). MC acknowledges support from the Swiss National Science Foundation (SNSF) (grants PA00P3_142120 and P300P3_154664). JG and OGJ acknowledge further funding support from USDA Hatch Act funds (IND0-1162). The authors declare no conflict of interest or competing interests.
References
- Charnley, M., Gilbert, J., Jones, O. G. and Reynolds, N. P. (2018). Preparation of amyloid fibril networks. Bio-protocol 8(4): e2733
- Dharmadana, D., Reynolds, N. P., Conn, C. E. and Valery, C. (2017). Molecular interactions of amyloid nanofibrils with biological aggregation modifiers: implications for cytotoxicity mechanisms and biomaterial design. Interface Focus 7(4): 20160160.
- Gilbert, J., Charnley, M., Cheng, C., Reynolds, N. P. and Jones, O. G. (2017a). Quantifying Young’s moduli of protein fibrils and particles with bimodal force spectroscopy. Biointerphases 12(4): 041001.
- Gilbert, J., Reynolds, N. P., Russell, S. M., Haylock, D., McArthur, S., Charnley, M. and Jones, O. G. (2017b). Chitosan-coated amyloid fibrils increase adipogenesis of mesenchymal stem cells. Mater Sci Eng C 79: 363-371.
- Jung, J. M., Savin, G., Pouzot, M., Schmitt, C. and Mezzenga, R. (2008). Structure of heat-induced beta-lactoglobulin aggregates and their complexes with sodium-dodecyl sulfate. Biomacromolecules 9(9): 2477-2486.
- Lara, C., Adamcik, J., Jordens, S. and Mezzenga, R. (2011). General self-assembly mechanism converting hydrolyzed globular proteins into giant multistranded amyloid ribbons. Biomacromolecules 12(5): 1868-1875.
- Reynolds, N. P., Adamcik, J., Berryman, J. T., Handschin, S., Zanjani, A. A. H., Li, W., Liu, K., Zhang, A. and Mezzenga, R. (2017). Competition between crystal and fibril formation in molecular mutations of amyloidogenic peptides. Nature Communications 8: 1338.
- Reynolds, N. P., Charnley, M., Bongiovanni, M. N., Hartley, P. G. and Gras, S. L. (2015). Biomimetic topography and chemistry control cell attachment to amyloid fibrils. Biomacromolecules 16(5): 1556-1565.
- Reynolds, N. P., Charnley, M., Mezzenga, R. and Hartley, P. G. (2014). Engineered lysozyme amyloid fibril networks support cellular growth and spreading. Biomacromolecules 15(2): 599-608.
- Reynolds, N. P., Styan, K. E., Easton, C. D., Li, Y., Waddington, L., Lara, C., Forsythe, J. S., Mezzenga, R., Hartley, P. G. and Muir, B. W. (2013). Nanotopographic surfaces with defined surface chemistries from amyloid fibril networks can control cell attachment. Biomacromolecules 14(7): 2305-2316.
- Usov, I. and Mezzenga, R. (2015). FiberApp: An open-source software for tracking and analyzing polymers, filaments, biomacromolecules, and fibrous objects. Macromolecules 48: 1269-1280.
- Wei, G., Su, Z., Reynolds, N. P., Arosio, P., Hamley, I. W., Gazit, E. and Mezzenga, R. (2017). Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. Chem Soc Rev 46(15): 4661-4708.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Charnley, M., Gilbert, J., Jones, O. G. and Reynolds, N. P. (2018). Characterization of Amyloid Fibril Networks by Atomic Force Microscopy. Bio-protocol 8(4): e2732. DOI: 10.21769/BioProtoc.2732.
Category
Biochemistry > Protein > Self-assembly
Cell Biology > Cell imaging > Atomic force microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










