- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Investigating Localization of Chimeric Transporter Proteins within Chloroplasts of Arabidopsis thaliana
Published: Vol 8, Iss 3, Feb 5, 2018 DOI: 10.21769/BioProtoc.2723 Views: 10107
Reviewed by: Dennis NürnbergSam-Geun KongAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2855 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1975 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1763 Views
Abstract
In this protocol, we describe a method to design chimeric proteins for specific targeting to the inner envelope membrane (IEM) of Arabidopsis chloroplasts and the confirmation of their localization by biochemical analysis. Specific targeting to the chloroplast IEM can be achieved by fusing the protein of interest with a transit peptide and an IEM targeting signal. This protocol makes it possible to investigate the localization of chimeric proteins in chloroplasts using a small number of transgenic plants by using a modified method of chloroplast isolation and fractionation. IEM localization of chimeric proteins can be further assessed by trypsin digestion and alkaline extraction. Here, the localization of the chimeric bicarbonate transporter, designated as SbtAII, is detected by Western blotting using antibodies against Staphylococcal protein A. This protocol is adapted from Uehara et al., 2016.
Keywords: AlkalineBackground
It has been proposed that the integration of cyanobacterial CO2 concentration mechanisms into chloroplasts is a promising approach to improve photosynthesis in C3 plants. According to theoretical estimations, integration of BicA and SbtA into the chloroplast IEM improves photosynthetic CO2 fixation rates. We examined the integration of nuclear-encoded cyanobacterial bicarbonate transporters, BicA and SbtA, to the IEM of chloroplasts in Arabidopsis. Therefore, we developed a protocol to design chimeric constructs for specific targeting of the IEM and investigate the localization of chimeric proteins in chloroplasts.
Materials and Reagents
- Construction of vectors and Arabidopsis transformation
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- 1.5 ml microtubes
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Arabidopsis chloroplast isolation
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- 1.5 ml microtubes
- Single-edge razor blades
- Plastic Petri plates, diameter 150 x 15 mm and diameter 90 x 15 mm
- 200 μm nylon mesh cone (Kyoshin Rikoh)
Note: 100 to 120 mm mesh squares were folded into a cone and stapled to hold its shape (Figure 1).
Figure 1. Procedure to make the 200 μm mesh cone - Protoplast-rupturing device (Figure 2)
- 10 ml disposable syringe (Terumo)
- 20 μm nylon mesh (Kyoshin Rikoh)
- 10 μm nylon mesh (Kyoshin Rikoh)
- Electrical tape, 15-20 mm wide
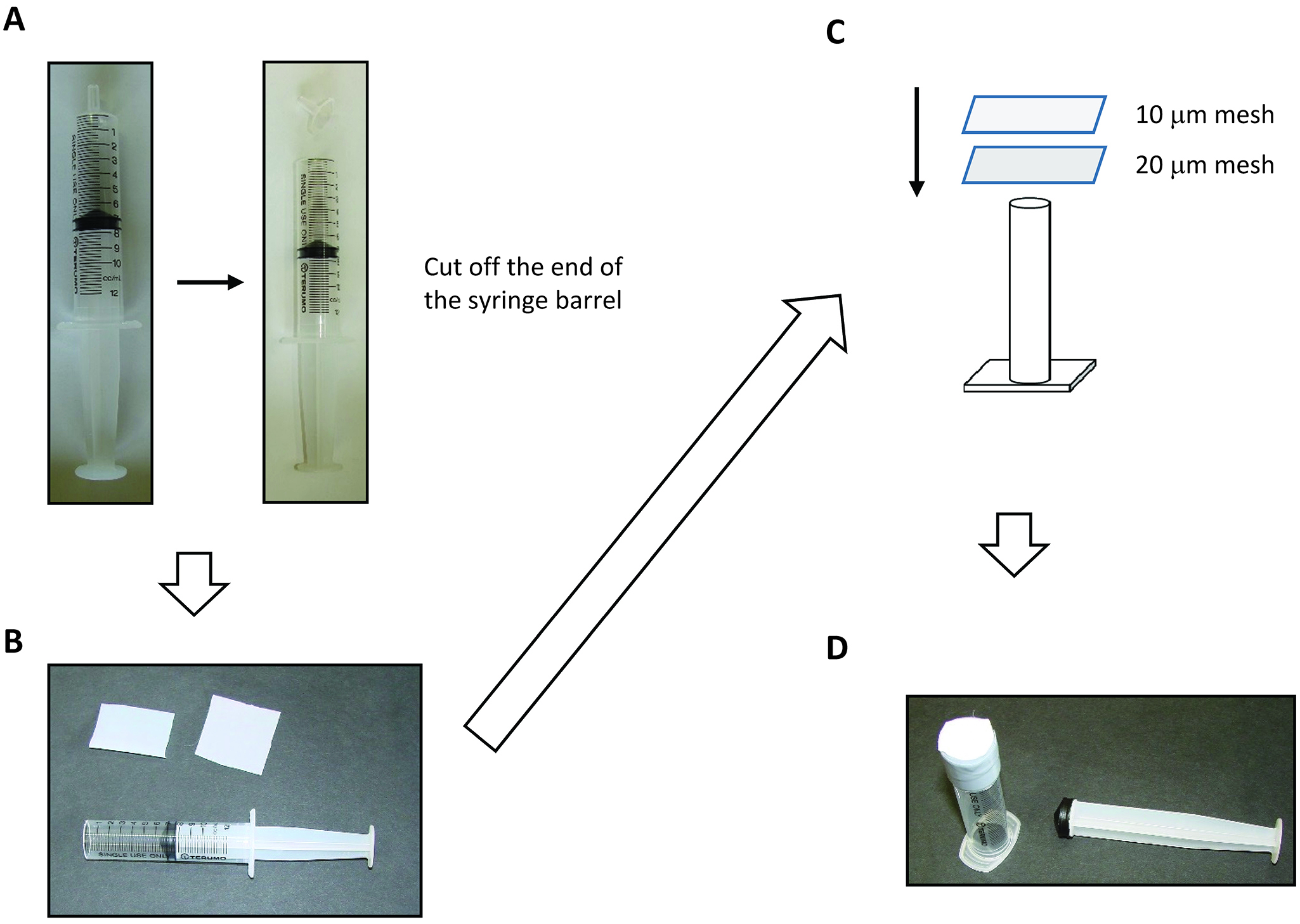
Figure 2. Procedure to make the protoplast-rupturing device. A. Cut off the end of syringe barrel. B. Syringe barrel, 20 μm mesh (Left), and 10 μm mesh. C. Schematic drawings of making the protoplast-rupturing device. D. Finished product of the protoplast-rupturing device. - 10 ml disposable syringe (Terumo)
- Pasteur pipet
- Arabidopsis (accession Columbia)
- Murashige and Skoog Plant salt mixture (Wako Pure Chemical Industries, catalog number: 392-00591 )
- Sucrose (Wako Pure Chemical Industries, catalog number: 196-00015 )
- 2-Morpholinoethanesulfonic acid, monohydrate (MES) (DOJINDO, catalog number: 343-01621 )
- Agar (Wako Pure Chemical Industries, catalog number: 016-11875 )
- Potassium hydroxide (KOH) (Wako Pure Chemical Industries, catalog number: 168-21815 )
- Sorbitol (Sigma-Aldrich, catalog number: S1876-1KG )
- Calcium chloride dihydrate (CaCl2·2H2O) (Wako Pure Chemical Industries, catalog number: 031-00435 )
- Cellulase (Yakult)
- Macerozyme (Yakult)
- Percoll (GE Healthcare Life Sciences, catalog number: 17089101 )
- 1 M 2-[4-(2-Hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES)-KOH, pH 7.5 and pH 8.0 (HEPES was purchased from DOJINDO, catalog number: 342-01375 )
Note: The pH of HEPES buffer was adjusted with KOH. - Magnesium chloride hexahydrate (MgCl2·6H2O) (Wako Pure Chemical Industries, catalog number: 135-00165 )
Note: 1 M magnesium chloride was used in Procedure B. - Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Wako Pure Chemical Industries, catalog number: 133-00725 )
Note: 1 M manganese(II) chloride was used in Procedure B. - Ethylenediaminetetraacetic acid (EDTA) (DOJINDO, catalog number: 345-01865 )
Note: 0.5 M EDTA was used in Procedure B. - Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3912-100G )
- Tricine (DOJINDO, catalog number: 347-02844 )
Note: Tricine was directly used in Procedure B. - O,O’-bis(2-aminoethyl)ethyleneglycol-N,N,N’,N’-tetraacetic acid (EGTA) (DOJINDO, catalog number: 346-01312 )
- Sodium hydrogen carbonate (NaHCO3) (Wako Pure Chemical Industries, catalog number: 191-01305 )
- 0.5x Murashige and Skoog medium (MS medium) (see Recipes)
- Digestion buffer (see Recipes)
- Digestion enzyme buffer (see Recipes)
- 40% (v/v) AT Percoll (see Recipes)
- 85% (v/v) AT Percoll (see Recipes)
- Gradient buffer (see Recipes)
- Protoplast resuspension buffer (see Recipes)
- Protoplast breakage buffer (see Recipes)
- HEPES-sorbitol buffer, pH 8.0 (see Recipes)
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Fractionation of chloroplasts
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- 1.5 ml microtubes
- Acetone (Wako Pure Chemical Industries, catalog number: 016-00346 )
Note: 80% (v/v) acetone was used in Procedure C. - 100% (w/v) trichloroacetic acid (TCA) (Wako Pure Chemical Industries, catalog number: 208-08081 )
- Sucrose (Wako Pure Chemical Industries, catalog number: 196-00015 )
- Potassium hydroxide (KOH) (Wako Pure Chemical Industries, catalog number: 168-21815 )
- Sorbitol (Sigma-Aldrich, catalog number: S1876-1KG )
- Ethylenediaminetetraacetic acid (EDTA) (DOJINDO, catalog number: 345-01865 )
Note: 0.5 M EDTA was used in Procedure C. - Tricine (DOJINDO, catalog number: 347-02844 )
Note: 1 M Tricine-KOH, pH 7.5, was made for TE/DTT buffer in Procedure C. - Dithiothreitol (DTT) (Wako Pure Chemical Industries, catalog number: 041-08976 )
Note: 1 M DTT was used in Procedure C. - Ribonucleic acid, transfer (tRNA) (MP Biomedicals, catalog number: 0215653480 )
- 2-Amino-2-hydroxymethyl-1,3-propanediol (Tris) base (Wako Pure Chemical Industries, catalog number: 207-06275 )
Note: 1 M Tris was used in Procedure C. - Sodium dodecyl sulfate (SDS) (Wako Pure Chemical Industries, catalog number: 196-08675 )
Note: 20% (w/v) SDS was used in Procedure C. - Glycerol (Wako Pure Chemical Industries, catalog number: 075-00616 )
Note: 50% (v/v) glycerol was used in Procedure C. - Saturated bromophenol blue (Wako Pure Chemical Industries, catalog number: 029-02912 )
- TE/DTT buffer (see Recipes) containing 1, 0.6, 0.46, 0.2, and 0 M sucrose
- SDS-sample buffer (see Recipes)
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Trypsin treatment of intact chloroplasts
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Sorbitol (Sigma-Aldrich, catalog number: S1876-1KG )
- Calcium chloride dihydrate (CaCl2·2H2O) (Wako Pure Chemical Industries, catalog number: 031-00435 )
Note: 1 M calcium chloride was used in Procedure D. - Percoll (GE Healthcare, catalog number: 17089101 )
- 1 M 2-[4-(2-Hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES)-KOH, pH 7.5 and pH 8.0 (HEPES was purchased from DOJINDO, catalog number: 342-01375 )
Note: The pH of 1 M HEPES buffer was adjusted with KOH. - Magnesium chloride hexahydrate (MgCl2·6H2O) (Wako Pure Chemical Industries, catalog number: 135-00165 )
Note: 1 M magnesium chloride was used in Procedure D. - Ethylenediaminetetraacetic acid (EDTA) (DOJINDO, catalog number: 345-01865 )
Note: 0.5 M EDTA was used in Procedure D. - Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3912-100G )
- Dithiothreitol (DTT) (Wako Pure Chemical Industries, catalog number: 041-08976 )
Note: 1 M DTT was used in Procedure D. - 2-Amino-2-hydroxymethyl-1,3-propanediol (Tris) base (Wako Pure Chemical Industries, catalog number: 207-06275 )
Note: 1 M Tris was used in Procedure D. - Sodium dodecyl sulfate (SDS) (Wako Pure Chemical Industries, catalog number: 196-08675 )
Note: 20 % (w/v) SDS was used in Procedure D. - Glycerol (Wako Pure Chemical Industries, catalog number: 075-00616 )
Note: 50% (v/v) glycerol was used in Procedure D. - Saturated bromophenol blue (Wako Pure Chemical Industries, catalog number: 029-02912 )
- Trypsin (Sigma-Aldrich, catalog number: T1005 )
Note: 20 mg/ml trypsin was used in Procedure D. - Nα-Tosyl-L-lysine chloromethyl ketone (TLCK) (Sigma-Aldrich, catalog number: T7254-100MG )
Note: 50 mg/ml TLCK was used in Procedure D. - Aprotinin (Sigma-Aldrich, catalog number: A3886-1VL )
Note: 2 mg/ml aprotinin was used in Procedure D. - Phenylmethanesulfonyl fluoride (PMSF) (Wako Pure Chemical Industries, catalog number: 164-12181 )
Note: 200 mM PMSF was used in Procedure D. - Trypsin inhibitor (Sigma-Aldrich, catalog number: T6522-25MG )
Note: 10 mg/ml trypsin inhibitor was used in Procedure D. - cOmpleteTM, EDTA-FREE (Roche Diagnostics, catalog number: 11 873 580 001 )
- 40% (v/v) AT Percoll (see Recipes)
- SDS-sample buffer (see Recipes)
- 2x trypsin buffer (see Recipes)
- 2x stop buffer (see Recipes)
- 1x 40% (v/v) Percoll (see Recipes)
- 1x HEPES-sorbitol buffer (see Recipes)
- SDS-cOmpleteTM buffer (see Recipes)
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Alkaline extraction of chloroplasts
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Acetone (Wako Pure Chemical Industries, catalog number: 016-00346 )
Note: 80 % (v/v) acetone was used in Procedure E. - 100% (w/v) trichloroacetic acid (TCA) (Wako Pure Chemical Industries, catalog number: 208-08081 )
- Dithiothreitol (DTT) (Wako Pure Chemical Industries, catalog number: 041-08976 )
Note: 1 M DTT was used in Procedure E. - Ribonucleic acid, transfer (tRNA) (MP Biomedicals, catalog number: 0215653480 )
- Sodium dodecyl sulfate (SDS) (Wako Pure Chemical Industries, catalog number: 196-08675 )
Note: 20 % (w/v) SDS was used in Procedure E. - Glycerol (Wako Pure Chemical Industries, catalog number: 075-00616 )
Note: 50% (v/v) glycerol was used in Procedure E. - Saturated bromophenol blue (Wako Pure Chemical Industries, catalog number: 029-02912 )
- Sodium carbonate (Na2CO3), pH 12 (Wako Pure Chemical Industries, catalog number: 199-01585 )
Note: 0.2 M sodium carbonate was used in Procedure E. - SDS-sample buffer (see Recipes)
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Dot blot assay for estimation of protein concentration
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- WhatmanTM 3MM Chromatography paper (GE Healthcare, catalog number: 3030-917 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3912-100G )
- Dithiothreitol (DTT) (Wako Pure Chemical Industries, catalog number: 041-08976 )
Note: 1 M DTT was used in Procedure F. - 2-Amino-2-hydroxymethyl-1,3-propanediol (Tris) base (Wako Pure Chemical Industries, catalog number: 207-06275 )
Note: 1 M Tris was used in Procedure F. - 20% (w/v) sodium dodecyl sulfate (SDS) (Wako Pure Chemical Industries, catalog number: 196-08675 )
- Glycerol (Wako Pure Chemical Industries, catalog number: 075-00616 )
Note: 50% (v/v) glycerol was used in Procedure F. - Saturated bromophenol blue (Wako Pure Chemical Industries, catalog number: 029-02912 )
- Coomassie Blue R-250 (Wako Pure Chemical Industries, catalog number: 031-17922 )
- Methanol (Wako Pure Chemical Industries, catalog number: 139-01827 )
- Acetic acid (Wako Pure Chemical Industries, catalog number: 017-00256 )
- Paper towel
- SDS-sample buffer (see Recipes)
- Coomassie Blue stain (see Recipes)
- Coomassie destain solution (see Recipes)
- Pipette tips (20 μl, 200 μl, 1,000 μl and 5 ml tips)
- Data analysis
- Antibody against the protein A (Sigma-Aldrich, catalog number: P3775 ) (this antibody was used for the detection of chimeric transporter protein tagged with protein A)
- Antibody against the large subunit (LSU) of Rubisco (a marker protein of the stroma)
- Antibody against Tic (Translocon at the inner envelope membrane of chloroplasts) 110 (a marker protein of the inner envelope membrane)
- Antibody against the light-harvesting complex protein (LHCP) (a marker protein of the thylakoid membrane)
- Antibody against Toc (Translocon at the outer envelope membrane of chloroplasts) 75 (a marker protein of the outer envelope membrane)
- Antibody against the protein A (Sigma-Aldrich, catalog number: P3775 ) (this antibody was used for the detection of chimeric transporter protein tagged with protein A)
Equipment
- Construction of vectors and Arabidopsis transformation
- Arabidopsis chloroplast isolation
- Pipettes (Gilson, models: P20, P200, P1000, and P5000, catalog numbers: F123600 , F123601 , F123602 , and F123603 )
- Refrigerator and Freezer
- Growth chamber (16 h light/8 h dark, 70-120 μE m-2 sec-1, 22 °C)
- 50 ml centrifuge tubes (IWAKI)
- Large-capacity centrifuge (TOMY SEIKO, model: Suprema 23 )
- Swinging-bucket rotor (TOMY SEIKO, models: TS-33N and B433 )
- Swinging-bucket (TOMY SEIKO, model: 3350-G01P )
- Pipettes (Gilson, models: P20, P200, P1000, and P5000, catalog numbers: F123600 , F123601 , F123602 , and F123603 )
- Fractionation of chloroplasts
- Pipettes (Gilson, models: P20, P200, P1000, and P5000, catalog numbers: F123600 , F123601 , F123602 , and F123603 )
- Refrigerator and Freezer
- Homogenizer (IUCHI)
- OptimaTM TL Ultracentrifuge (Beckman Coulter)
- Angle rotor (Beckman Coulter, model: TLA-100.3 , catalog number: 349490)
- 3.5 ml polycarbonate ultracentrifuge tubes (Beckman Coulter, catalog number: 349622 )
- Swinging-bucket rotor (Beckman Coulter, model: TLS-55 , catalog number: 346134)
- 2.2 ml Ultra-ClearTM centrifuge tubes (Beckman Coulter, catalog number: 347356 )
- Pipettes (Gilson, models: P20, P200, P1000, and P5000, catalog numbers: F123600 , F123601 , F123602 , and F123603 )
- Trypsin treatment of intact chloroplasts
- Alkaline extraction of chloroplasts
- Pipettes (Gilson, models: P20, P200, P1000, and P5000, catalog numbers: F123600 , F123601 , F123602 , and F123603 )
- Angle rotor (Beckman Coulter, model: TLA-100.3 , catalog number: 349490)
- OptimaTM TL Ultracentrifuge (Beckman Coulter)
- 1.5 ml Polyallomer tubes (Beckman Coulter, catalog numbers: 357448 , 355919 )
- Pipettes (Gilson, models: P20, P200, P1000, and P5000, catalog numbers: F123600 , F123601 , F123602 , and F123603 )
- Dot blot assay for estimation of protein concentrations
Procedure
- Construction of vectors and Arabidopsis transformation
To deliver a protein of interest to the chloroplast IEM, we have to fuse at least two distinct targeting signals to it. One signal is the transit peptide, and the other is the chloroplast IEM targeting signal (Figure 3). The transit peptide has been known to serve as the targeting signal to the chloroplast stroma and found in the majority of precursor proteins targeted to the chloroplast interior. However, the transit peptide seems to be insufficient to target cargo proteins, in our case the cyanobacterial bicarbonate transporters, to the chloroplast IEM. As an additional IEM targeting signal, we fused the mature portion of the Cor413im1 protein to the chimeric construct shown in Figure 3. This portion can function as the IEM targeting signal in transgenic Arabidopsis (Uehara et al., 2016). Another study has shown that a membrane protein leader can serve as the IEM targeting signal in a transient expression system (Rolland et al., 2016). In addition, it is necessary to add a tag to detect chimeric proteins if specific antibodies are unavailable.
The length of the transit peptide and the mature portion of Cor413im1 shown in Figure 3 has been already described in detail elsewhere (Okawa et al., 2008 and 2014).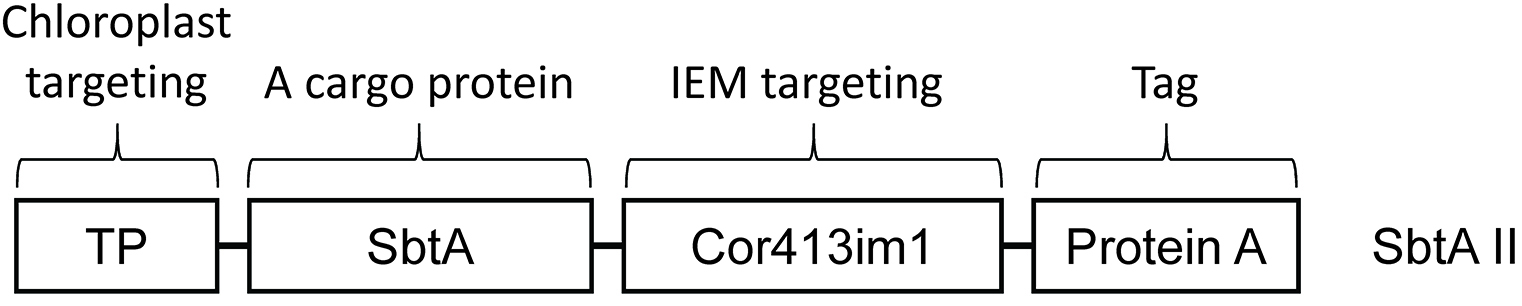
Figure 3. Construct design for the specific targeting of bicarbonate transporter SbtA II to the chloroplast IEM. Schematic diagram of the chimeric SbtAII construct used in this study. TP indicates transit peptide and serves as the targeting signal to chloroplasts. The Cor413im1 portion of the construct was derived from the mature portion of Cor413im1 and can function as the targeting signal to the chloroplast IEM. The protein A domain of the fusion construct contains two IgG-binding domains from Staphylococcal protein A and serves as the tag for detection of the chimeric protein. - Arabidopsis chloroplast isolation
We developed a method to isolate sufficient amounts of intact chloroplasts using a smaller number of plants based on the method described in Smith et al. (2002). This method is useful if large numbers of seeds cannot be obtained for the isolation of chloroplasts (e.g., mutants, transgenic lines). Throughout the procedure, samples should be kept on ice unless stated otherwise, and buffers should be chilled prior to experiments.- Sow sterilized seeds on 0.5x MS plates (100 seeds per plate, four plates per isolation, see Recipes) supplemented with 1% sucrose. Allow Arabidopsis plants to grow in growth chambers under a 16 h-light/8 h-dark cycle, 100 μE m-2 sec-1, 22 °C for 14-18 d (Figure 4A).
- Harvest the entire aerial portions of the plants with a razor blade, and place immediately in a 90 mm diameter Petri dish containing 10 ml digestion buffer (see Recipes) on ice. Plants harvested from two plates should be placed into one enzyme digestion reaction so that you need to prepare two 90 mm diameter Petri dishes for four Arabidopsis plates. When all tissue has been harvested, chop the tissue rapidly using a razor blade for up to 1 min (Figure 4B).
Note: Do not chop for more than 1 min or smash the tissues. - Remove the digestion buffer from the Petri dish. Add 13.5 ml digestion enzyme solution (see Recipes) to the Petri dish, and distribute the tissue uniformly with the fingertips (Figure 4C). Place the Petri dishes into a plastic container, and incubate in a growth chamber for 3 h at 22 °C (light intensity at around 50-100 μE m-2 sec-1).
- During digestion, prepare a 40% (20 ml, upper layer):85% (7 ml, lower layer) AT Percoll (see Recipes) step gradient in a 50 ml centrifuge tube and maintain on ice. We use one gradient for two Arabidopsis plates (one enzyme digestion) so that we usually prepare two gradients for one isolation. You may also use smaller Percoll volumes and tubes (Smith et al., 2002).
- Harvest the protoplasts through gentle swirling (or gentle agitation) of the plate for 30-60 sec, then filter the medium through a 200 μm mesh cone, placed inside a small funnel, into a 50 ml centrifuge tube on ice. Collect protoplasts from one Petri dish into one 50 ml tube that you need to prepare two tubes. Transfer any tissue from the nylon mesh back into the Petri dish and wash the tissue with 10 ml of ice-cold fresh digestion buffer. Filter the tissue-buffer mixture into the same 50 ml centrifuge tube. Wash the tissue one or two times more to ensure that as many protoplasts are released as possible (Figure 4D).
- Centrifuge the protoplasts for 5 min at 100 x g, 4 °C (Figure 4E). Remove and discard the supernatant carefully with an aspirator. Be careful not to disturb the protoplast pellet, as it is very loose.
- Resuspend the pellet in 5 ml protoplast resuspension buffer (see Recipes) by gently swirling the pellet while the buffer is dispensed down the side of the tube. After the protoplasts are resuspended, add another 5 ml protoplast resuspension buffer and gently mix (Figure 4F). Centrifuge the protoplasts for 4 min at 100 x g, 4 °C (Figure 4G).
- Remove the supernatant and resuspend the pellet in 5 ml protoplast breakage buffer (see Recipes). After the protoplasts are resuspended, add another 5 ml protoplast breakage buffer and gently mix (Figure 4H). Immediately transfer the resuspended protoplast pellet into the barrel of the protoplast-rupturing device. Holding the end of the device over a 50 ml centrifuge tube on ice, carefully replace the plunger, and gently and firmly force the suspension through the layers of mesh (Figure 4I). Repeat this procedure once.
- Quickly and carefully layer the 10 ml broken protoplasts onto the AT Percoll step gradient (Figure 4J). Centrifuge in a swinging-bucket rotor for 10 min at 2,500 x g, 4 °C, with the brake off (Figure 4K). Following centrifugation, there should be two visible green bands in the gradient: an upper band of broken chloroplasts at the protoplast breakage buffer/40% Percoll interface, and a lower band of intact chloroplasts at 40%/85% Percoll interface. Remove the load zone and the upper band of broken chloroplasts by aspiration. Harvest the lower band using a Pasteur pipet, and dilute the intact chloroplasts with 40-45 ml HEPES-sorbitol buffer (see Recipes), pH 8.0, in a 50 ml centrifuge tube (Figure 4L). Intact chloroplasts from the two AT Percoll gradients should be combined into one tube.
- Centrifuge the diluted intact chloroplasts for 5 min at 700 x g, 4 °C (Figure 4M). Carefully decant the supernatant, and discard all excess buffer without disrupting the pellet. Resuspend the pellet in a small volume (200-300 μl) of HEPES-sorbitol buffer, pH 8.0 (Figure 4N).
- Dilute 5 μl of the chloroplast resuspension into 995 μl 80% acetone. Mix vigorously and microcentrifuge at ~15,000 x g for 2 min to remove the protein precipitate. Measure the A652 of the supernatant against an 80% acetone blank. Calculate the chlorophyll concentration (mg/ml) by using the following equation (Smith et al., 2002):
(A652/36) x 200
to compensate for the dilution factor. Adjust the chlorophyll concentration to 1 mg chlorophyll/ml using HEPES-sorbitol buffer. The prepared chloroplasts should be used immediately for Procedures C and D, while chloroplasts for procedure E can be stored at -80 °C.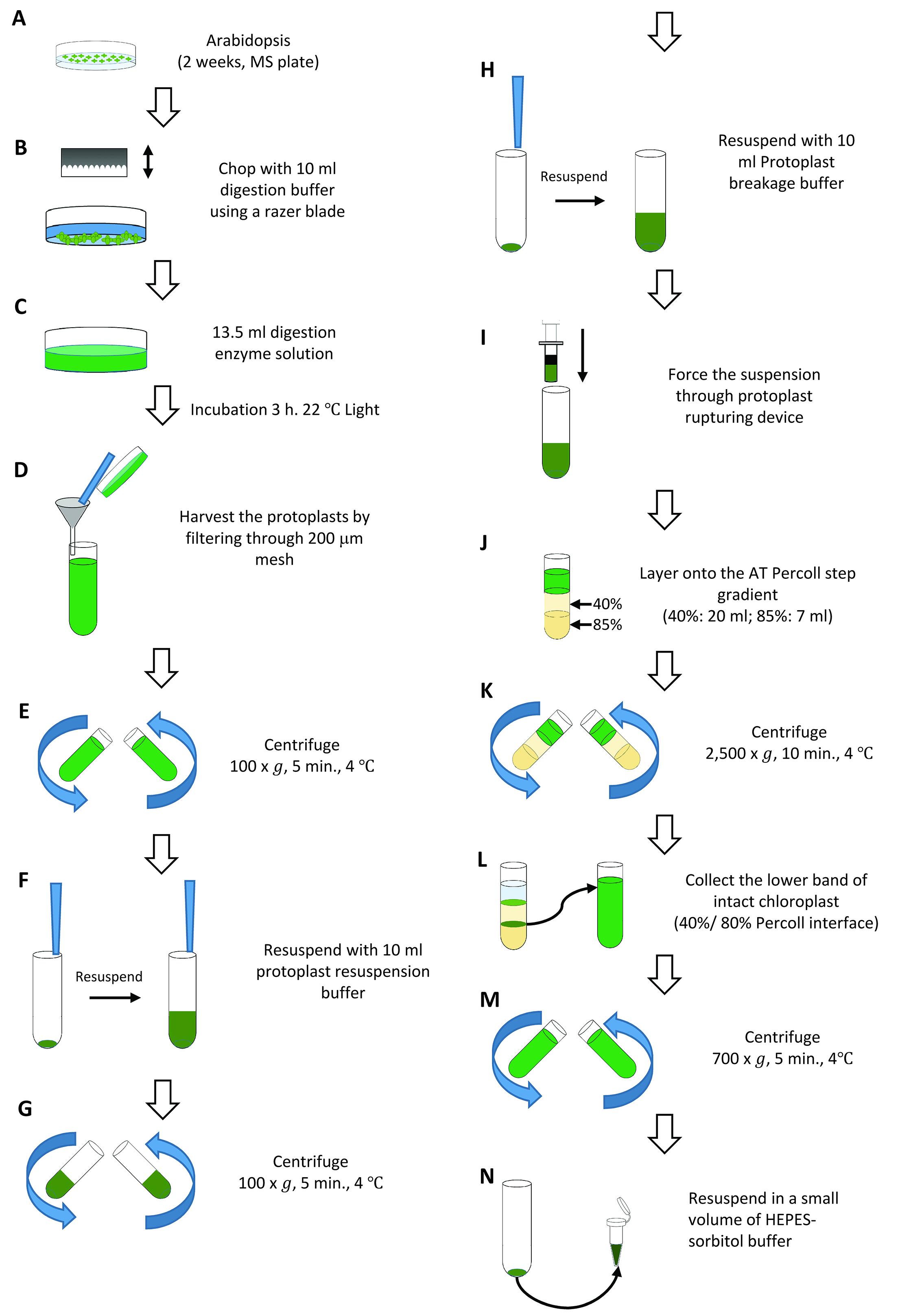
Figure 4. Overview of Arabidopsis chloroplasts isolation (Procedure B)
- Sow sterilized seeds on 0.5x MS plates (100 seeds per plate, four plates per isolation, see Recipes) supplemented with 1% sucrose. Allow Arabidopsis plants to grow in growth chambers under a 16 h-light/8 h-dark cycle, 100 μE m-2 sec-1, 22 °C for 14-18 d (Figure 4A).
- Fractionation of chloroplasts
- Centrifuge 0.5 ml of 1 mg chlorophyll/ml chloroplasts prepared in Step B11 for 5 min at 700 x g, 4 °C. Remove the supernatant and measure the volume of supernatant by pipetting. Resuspend the chloroplast pellet in TE/DTT buffer (see Recipes) containing 0.6 M sucrose to a concentration of 1 mg/ml chlorophyll (Figure 5A). Let stand for 10 min on ice. Freeze the chloroplast suspension for 1-2 h at -20 °C. If you do not proceed to the fractionation stage immediately, it is possible to keep the frozen chloroplast suspension at -80 °C until use.
- Thaw the suspension and dilute with 3 volumes of TE/DTT buffer. To collect the total chloroplast proteins, take 5 μl of chloroplast suspension into 1.5 ml microtubes before the addition of TE/DTT. Homogenize with 20 strokes in a Dounce (or Potter) homogenizer with a tight pestle (Figure 5B). Transfer the suspension into 3.5 ml polycarbonate ultracentrifuge tubes. Ultracentrifuge the lysed chloroplasts in a fixed angle rotor for 1 h at 40,000 x g, 4 °C (Figure 5C).
- Remove the brownish supernatant containing the stromal content using a pipette and store at -80 °C (Figure 5D). Resuspend the membrane pellet in TE/DTT buffer containing 0.2 M sucrose to a concentration of 1 mg chlorophyll/ml by pipetting (Figure 5D) and homogenization (Figure 5E, 20 strokes in a Dounce homogenizer).
- Set up a sucrose step gradient consisting of 1 ml of 1 M sucrose, and 0.7 ml of 0.46 M sucrose in 2.2 ml polyallomer tubes. Layer the membrane suspension (0.2 M sucrose) onto the top (Figure 5F and Figure 6A). Apply 0.5 ml of membrane suspension on each gradient. Ultracentrifuge the samples for 1.5 h at 270,000 x g, 4 °C, in a swinging-bucket rotor using low acceleration and deceleration rates (Figure 5G). Collect the interface from each step of the gradient (Figure 5H and Figure 6B). The upper interface (0.2:0.46 M sucrose) contains residual stroma and plastoglobules. The middle interface (0.46:1 M sucrose) is highly enriched in envelope membranes. The thylakoid membranes form a tight pellet at the bottom of the tube.
- Precipitate the stromal proteins (collected in Step C2) by adding 100% trichloroacetic acid (TCA) to give a final concentration of 10% and tRNA to give a final concentration of 10 μg/ml. Incubate for 60 min or longer on ice. Collect the TCA precipitate by centrifugation for 30 min at ~20,000 x g, 4 °C. Wash the pellet with 1 ml of 80% acetone and centrifuge for 15 min at ~20,000 x g, 4 °C. Resuspend the pellet directly in 200 μl SDS-sample buffer (see Recipes).
- Precipitate the total chloroplast proteins (5 μl collected in Step C2) by adding 1 ml of 80% acetone and vortex. Keep the tube on ice for 30 min and centrifuge for 30 min at ~20,000 x g, 4 °C. Resuspend the pellet directly in 50 μl SDS-sample buffer.
- To collect the envelope membranes, dilute the 0.46:1 M sucrose interface fraction with 3 to 5 volumes of TE/DTT buffer and centrifuge in a fixed angle rotor for 1 h at 270,000 x g, 4 °C (Figure 5J). Remove the supernatant with a pipette and discard. Resuspend the pellets in 30 μl of SDS-sample buffer for analysis (Figure 5K).
- Thylakoid pellets are suspended in 0.3 ml SDS-sample buffer (Figure 5H).
- After the quantification of proteins in each fraction, analyze total chloroplast (3 μg), stroma (3 μg), envelope (1 μg), and thylakoid (1.5 μg) fractions by SDS-PAGE.
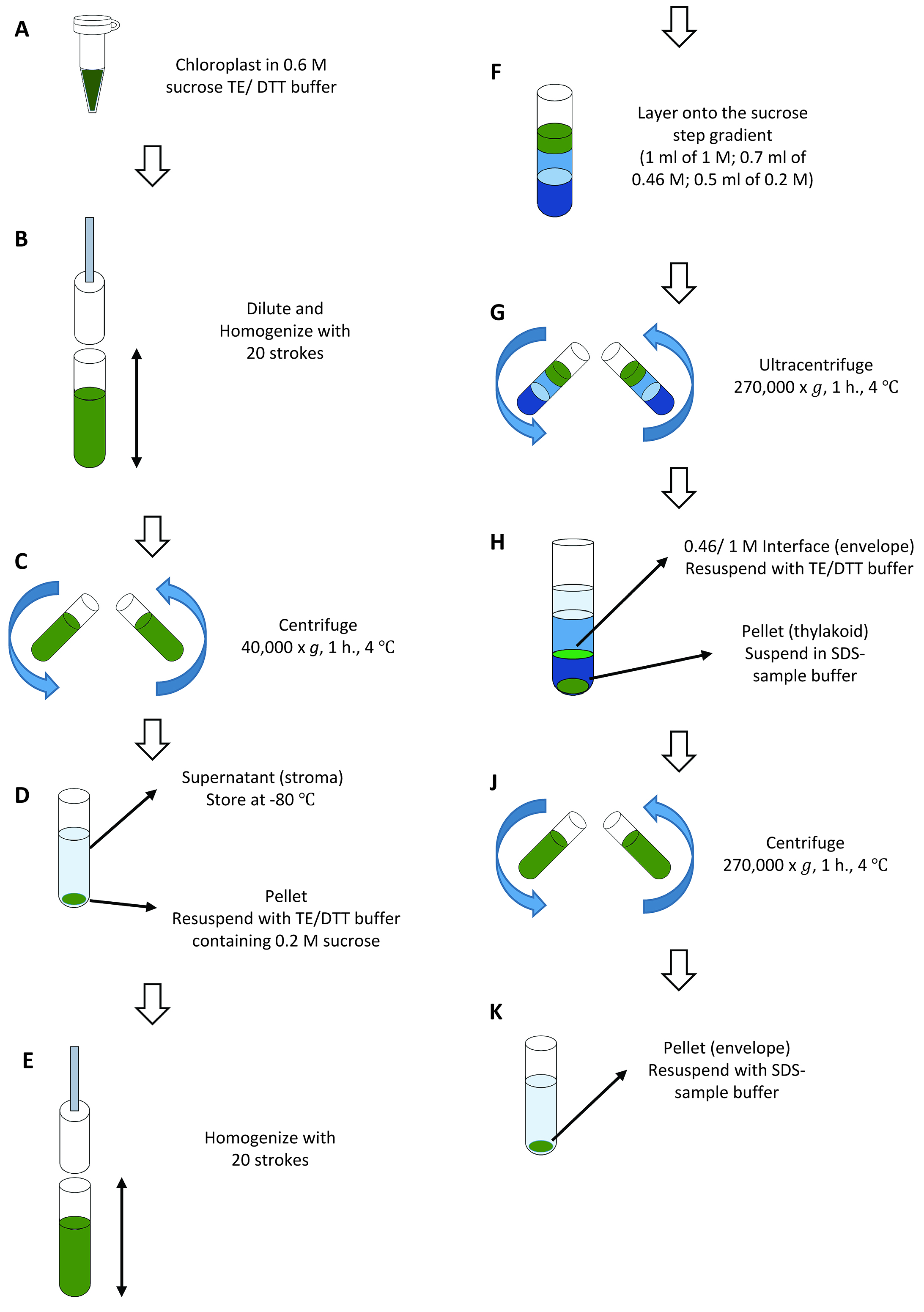
Figure 5. Overview of chloroplastfractionation (Procedure C)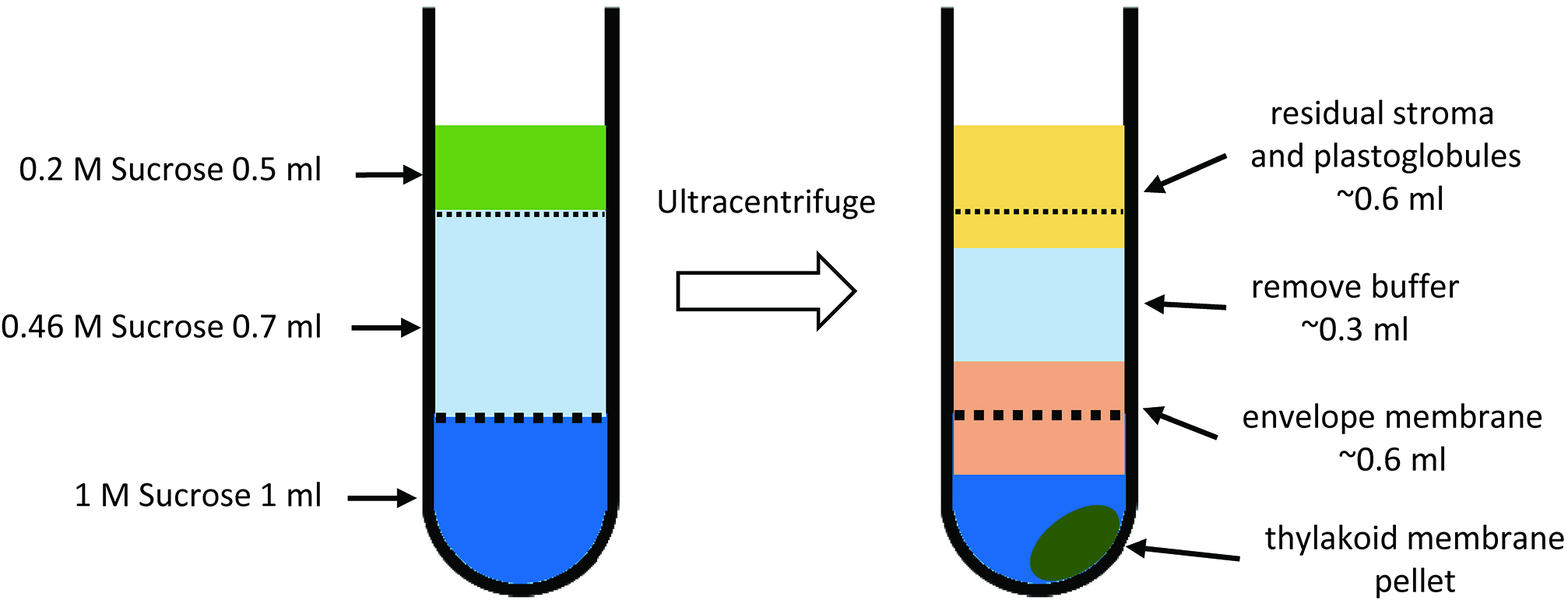
Figure 6. Schematic drawings of sucrose density gradient ultracentrifugation for the separation of envelope and thylakoid membrane (Step C4). This experiment uses 2.2 ml Ultra-ClearTM centrifuge tubes. A and B represent sucrose density gradients before and after ultracentrifugation, respectively.
- Centrifuge 0.5 ml of 1 mg chlorophyll/ml chloroplasts prepared in Step B11 for 5 min at 700 x g, 4 °C. Remove the supernatant and measure the volume of supernatant by pipetting. Resuspend the chloroplast pellet in TE/DTT buffer (see Recipes) containing 0.6 M sucrose to a concentration of 1 mg/ml chlorophyll (Figure 5A). Let stand for 10 min on ice. Freeze the chloroplast suspension for 1-2 h at -20 °C. If you do not proceed to the fractionation stage immediately, it is possible to keep the frozen chloroplast suspension at -80 °C until use.
- Trypsin treatment of intact chloroplasts
Trypsin is capable of permeating the outer envelope membrane (OEM) but not the inner envelope membrane (IEM) of intact chloroplasts (Jackson et al., 1998; Inaba et al., 2003). Therefore, proteins localized to the OEM, such as Toc75, are sensitive to trypsin treatment. This method allows us to determine whether a protein of interest localizes to the OEM or IEM of chloroplasts.- Dilute 25 μl of 1 mg chlorophyll/ml intact chloroplasts (Step B11) with 125 μl of HEPES-sorbitol buffer.
- Add 150 μl of 2x trypsin buffer (see Recipes), mix gently, and incubate for 30-40 min on ice.
- Stop the reaction by adding 300 μl of 2x stop buffer (see Recipes). Mix very carefully by inverting the tube several times and incubate on ice for 10 min. After incubation on ice, isolate the chloroplasts by centrifugation through a 1x 40% Percoll (see Recipes) for 5 min at 2,500 x g, 4 °C.
- Carefully aspirate the supernatant and 1x 40% Percoll, containing some broken chloroplasts.
- Resuspend the chloroplast pellet in 500 μl of 1x HEPES-sorbitol buffer (see Recipes). Centrifuge the chloroplasts for 2 min at 2,500 x g, 4 °C. Remove the supernatant and resuspend the pellet in 50 μl of SDS-cOmpleteTM buffer (see Recipes).
- Dilute 25 μl of 1 mg chlorophyll/ml intact chloroplasts (Step B11) with 125 μl of HEPES-sorbitol buffer.
- Alkaline extraction of chloroplasts
To investigate whether a protein of interest is integrated into the envelope membranes, chloroplasts can be subjected to alkaline extraction. If the protein is integrated into the envelope membranes, it should be recovered in the pellet fraction after alkaline extraction and subsequent ultracentrifugation. Peripherally associated IEM proteins, such as Tic22, are released from the IEM during the extraction (Kouranov et al., 1998).- Dilute 25 μl of 1 mg chlorophyll/ml chloroplasts (Step B11) into 1 ml of 0.2 M Na2CO3, pH 12, in a 1.5 ml polyallomer tube and incubate for 10 min on ice.
- Ultracentrifuge the mixture in a fixed angle rotor for 15 min at 100,000 x g, 4 °C. Carefully collect the supernatant containing the stroma and transfer to a fresh tube. Resuspend the membrane pellet in 80 μl SDS-sample buffer. Dissolve the pellet completely.
- Precipitate the alkaline supernatant by adding 100% (w/v) TCA to a final concentration of 10% (w/v) and tRNA to a final concentration of 10 μg/ml. Incubate for 30 to 60 min on ice.
- Collect the TCA precipitate by centrifuging for 30 min at ~20,000 x g, 4 °C. Wash the pellet with 1 ml of 80% acetone and centrifuge for 15 min at ~20,000 x g, 4 °C.
- Resuspend the pellet directly in 80 μl SDS-sample buffer. Dissolve the pellet completely.
- Dilute 25 μl of 1 mg chlorophyll/ml chloroplasts (Step B11) into 1 ml of 0.2 M Na2CO3, pH 12, in a 1.5 ml polyallomer tube and incubate for 10 min on ice.
- Dot blot assay for estimation of protein concentrations
Estimation of concentrations of samples from Procedures C and D are performed as follows:- Cut a square of WhatmanTM 3MM Chromatography paper (3MM paper) and place it on a clean sheet of paper towel.
- Mark the concentration of BSA standards and each sample to be tested on 3MM paper with a pencil (see Figure 7).
- Dilute the 2.0 mg/ml bovine serum albumin (BSA) with SDS-sample buffer to make BSA standards (0.2, 0.4, 0.6, 0.8, 1.0, 1.2, and 1.5 mg/ml). Dilute 3 μl of each sample to be tested with 3 μl SDS-sample buffer. If the protein concentration is expected to be high, make a serial dilution as shown in Figure 7.
- Gently dot 2 μl of the BSA standards and each sample onto the 3MM paper with a pipette.
- Dry the 3MM paper at room temperature for at least 20 min.
- Stain on a shaker with Coomassie Blue stain for 30 min.
- Collect Coomassie Blue stain as much as possible (reusable).
- Destain the paper on a shaker with Coomassie destain solution. Change the destain solution every 20 min. Destain until the background is clear.
- Dry the 3MM paper on a paper towel.
- Estimate the concentration of each sample by comparing the intensity of the blot color to the BSA standards.
- Cut a square of WhatmanTM 3MM Chromatography paper (3MM paper) and place it on a clean sheet of paper towel.
Data analysis
Determination of protein concentration and SDS-PAGE
- Compare the strength of each signal to the BSA standards and determine the concentration of protein (Figure 7: e.g., the signal strength of the 1/2-diluted Cp spot is almost equivalent to that of the 0.6 mg/ml BSA standard spot).
- Prepare samples based on quantification of proteins in each fraction. Sample volumes can be adjusted using SDS-sample buffer.
Note: In most cases, a portion of each fraction is sufficient to detect the protein of interest. The protein ratio of Cp:Str:Env:Thy (prepared in Procedure C) should be consistently 3:3:1:1.5.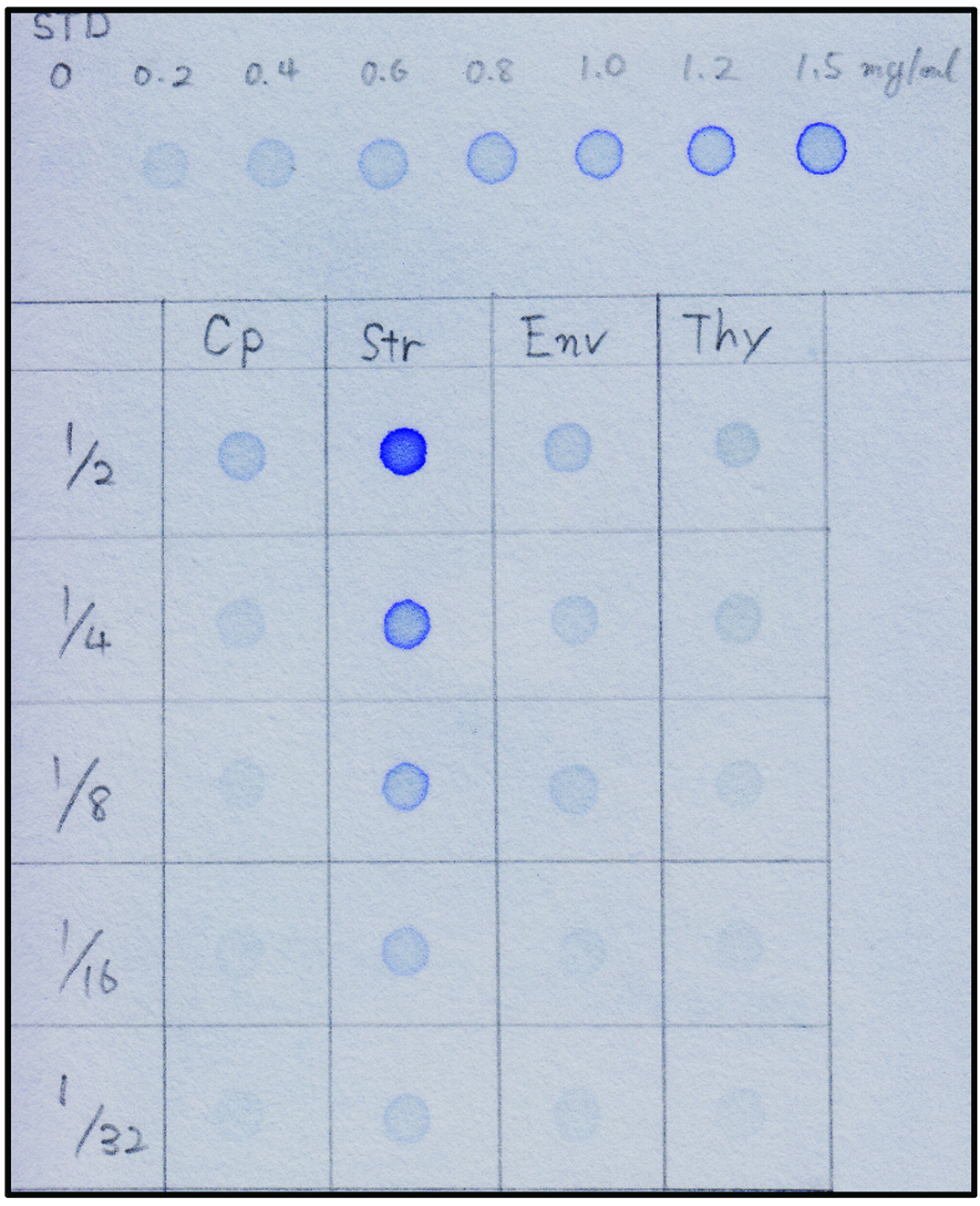
Figure 7. Dot blot quantification of protein concentration. The upper portion indicates the staining of BSA standards (STD). Isolated chloroplasts (Cp) were fractionated into stroma (Str), envelope membrane (Env), and thylakoid membrane (Thy) fractions in Procedure C. Each sample was diluted 2, 4, 8, 16, 32 times with SDS-sample buffer. - The proteins recovered in each fraction (Procedures C, D, and E) were resolved by SDS-PAGE and analyzed by Western blotting.
- Investigate the localization of chimeric proteins within the chloroplasts. As an example, we used SbtAII. The purity of each fraction was confirmed using marker proteins such as LSU (stroma), Tic110 (envelope membrane), and LHCP (thylakoid membrane). The SbtAII fusion protein was localized in chloroplasts (Figure 8, lane Cp). Furthermore, the SbtAII fusion protein was found to be highly enriched in the envelope fraction (Figure 8, lane Env).
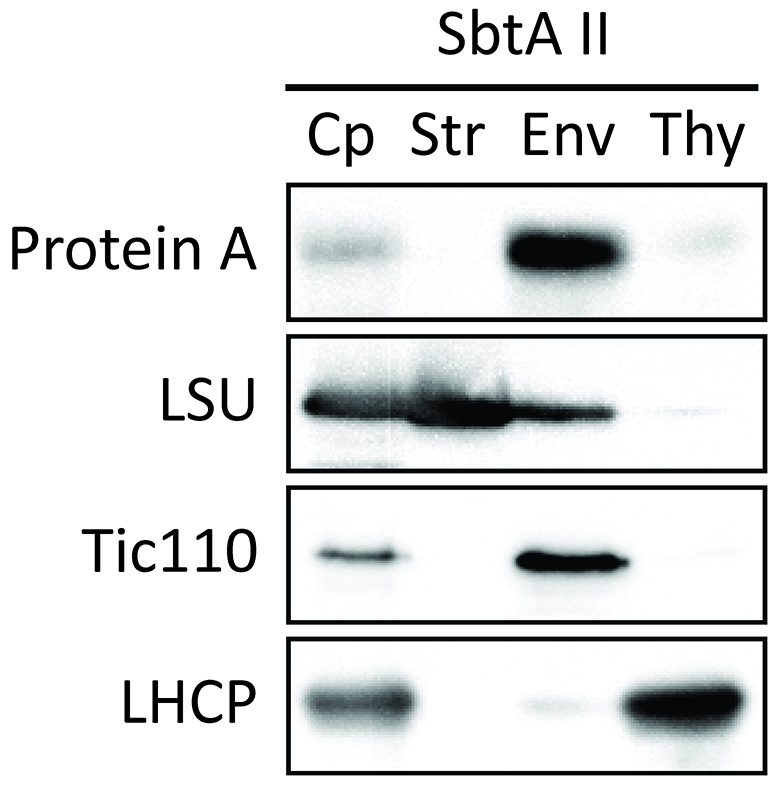
Figure 8. Localization of chimeric protein SbtAII in Arabidopsis chloroplasts (Procedure C). Isolated chloroplasts (Cp) were fractionated into stroma (Str), envelope membrane (Env), and thylakoid membrane (Thy) fractions. The protein ratio of Cp:Str:Env:Thy used in these analyses was consistently 3:3:1:1.5. Each fraction was western blotted with antibodies against protein A, LSU, Tic110 and LHCP. - Investigate whether SbtAII is an outer or inner envelope membrane protein. Trypsin permeates the outer envelope membrane, but not the inner envelope membrane, of intact chloroplasts. As expected, the OEM protein Toc75 was digested by trypsin (Figure 9, Toc75). In contrast, chimeric SbtAII (Figure 9, Protein A) and the IEM protein, Tic110 (Figure 9, Tic110), are resistant to trypsin, indicating that the chimeric protein is localized to the IEM of chloroplasts.
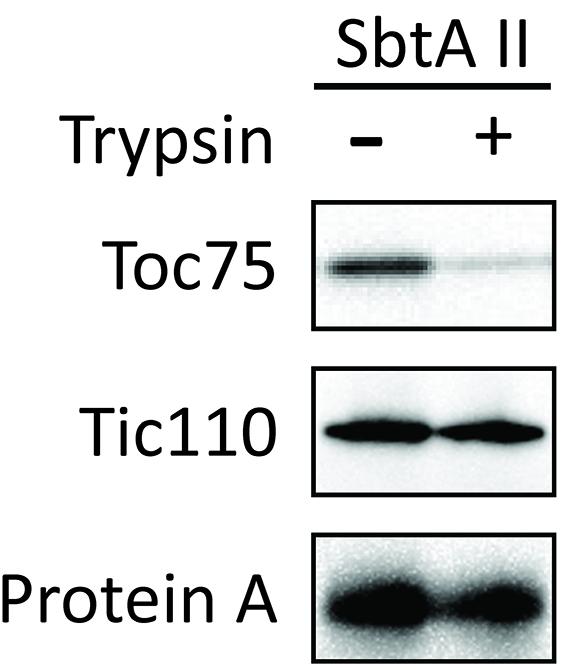
Figure 9. Trypsin sensitivity of chimeric SbtA protein in chloroplasts (Procedure D). The protease sensitivity of the outer envelope membrane protein, Toc75, was included as a positive control. The protease resistance of the inner envelope membrane protein, Tic110, was also included as a control. - Investigate whether SbtAII is integrated into the IEM, or is peripherally associated with the IEM. Tic110 is an integral IEM protein and is shown as a positive control. SbtAII was resistant to alkaline extraction (Figure 10), indicating that SbtAII is an integral membrane protein in the chloroplast IEM.
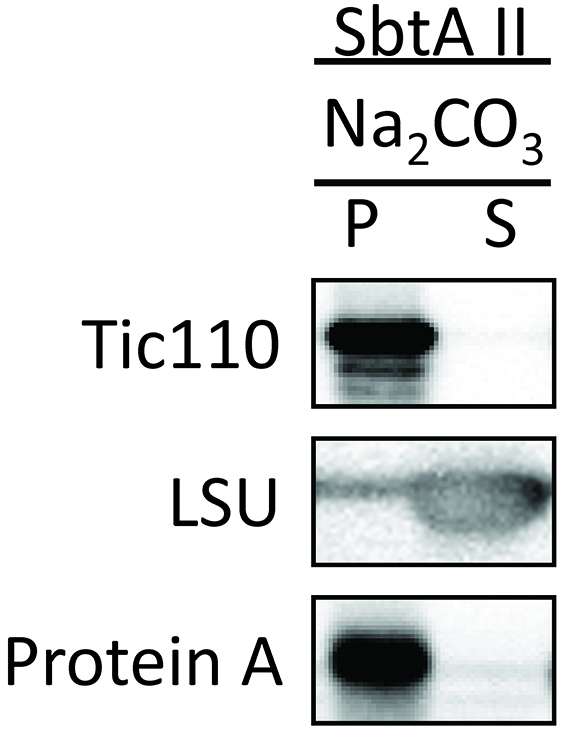
Figure 10. Localization of chimeric SbtA II protein in the soluble and membrane fractions of chloroplasts (Procedure E). Chloroplasts treated with alkaline solution were separated into insoluble (P) and soluble (S) fractions. The insoluble protein, Tic110, and the soluble protein, LSU, are included as controls.
Recipes
- 0.5x Murashige and Skoog medium (MS medium) (60 ml per plate)
0.23 g Murashige and Skoog Plant salt mixture (final 0.5x)
1 g sucrose (final 1% (w/v))
0.05 g MES (final 2.3 mM)
ddH2O to 100 ml, adjust pH to 5.7-5.8 using 1 N KOH
After adjusting, add 0.5 g agar (final 0.5%) and autoclave - Digestion buffer (store up to 2 weeks at 4 °C)
1.56 g MES (final 20 mM)
29.13 g sorbitol (final 400 mM)
200 μl 1 M CaCl2 (final 0.5 mM)
ddH2O to 400 ml, adjust pH to 5.2 using 1 N KOH - Digestion enzyme buffer
Dissolve 0.6 g cellulase and 0.12 g macerozyme in 30 ml digestion buffer
Centrifuge for 10 min at 2,000 x g to pellet insoluble materials
Use supernatant immediately - 40% (v/v) AT Percoll (store up to 1 month at -20 °C, thaw immediately before use)
40 ml Percoll (final 40% (v/v))
50 ml gradient buffer (final 50% (v/v))
10 ml ddH2O - 85% (v/v) AT Percoll (store up to 1 month at -20 °C, thaw immediately before use)
42.5 ml Percoll (final 85% (v/v))
2.5 ml 1 M HEPES-KOH, pH 7.5 (final 50 mM)
3.01 g sorbitol (final 330 mM)
Add ddH2O to 50 ml - Gradient buffer (store up to 2 weeks at 4 °C)
10 ml 1 M HEPES-KOH, pH 7.5 (final 100 mM)
12.04 g sorbitol (final 660 mM)
200 μl 1 M MgCl2 (final 2 mM)
200 μl 1 M MnCl2 (final 2 mM)
800 μl 0.5 M EDTA (final 4 mM)
0.2 g BSA (final 0.2% (w/v)) (add immediately before use)
ddH2O to 100 ml - Protoplast resuspension buffer (store up to 2 weeks at 4 °C)
0.39 g MES (final 20 mM)
7.29 g sorbitol (final 400 mM)
50 μl 1 M CaCl2 (final 0.5 mM)
ddH2O to 100 ml, adjust pH to 6.0 using 1 N KOH - Protoplast breakage buffer (store up to 2 weeks at 4 °C)
0.36 g Tricine (final 20 mM)
5.47 g sorbitol (final 300 mM)
1 ml 0.5 M EDTA (final 5 mM)
0.19 g EGTA (final 5 mM)
0.084 g NaHCO3 (final 10 mM)
0.1 g BSA (final 0.1% (w/v)) (add immediately before use)
ddH2O to 100 ml, adjust pH to 8.4 using 1 N KOH - HEPES-sorbitol buffer, pH 8.0 (store up to 2 weeks at 4 °C)
15 ml 1 M HEPES-KOH, pH 8.0 (final 50 mM)
18.06 g sorbitol (final 330 mM)
ddH2O to 300 ml - TE/DTT buffer containing 1, 0.6, 0.46, 0.2 and 0 M sucrose (store up to 2 weeks at 4 °C)
0.25 ml 1 M Tricine-KOH, pH 7.5 (final 50 mM)
20 μl 0.5 M EDTA (final 2 mM)
5 μl 1 M DTT (final 1 mM) (add immediately before use)
1.712, 1.046, 0.787, 0.343 g sucrose (final 1, 0.6, 0.46, 0.2, and 0 M)
ddH2O to 5 ml - SDS-sample buffer (store at -20 °C until used)
0.35 ml 1 M Tris base (final 350 mM)
0.25 ml 20% (w/v) SDS (final 5% (w/v))
80 μl 1 M DTT (final 80 mM)
0.15 ml 50% (v/v) glycerol (final 7.5% (v/v))
40 μl saturated bromophenol blue (final 1% (v/v))
0.13 ml ddH2O - 2x trypsin buffer
8 μl 1 M MgCl2 (final 8 mM)
0.2 μl 1 M CaCl2 (final 0.2 mM)
HEPES-sorbitol buffer to 1 ml
Add 6.6 μl 20 mg/ml trypsin in 300 μl 2x trypsin buffer - 2x stop buffer
Add 2 μl 50 mg/ml TLCK
2 μl 2 mg/ml aprotinin
10 μl 200 mM PMSF
20 μl 10 mg/ml trypsin inhibitor
20 μl 0.5 M EDTA
in 1 ml HEPES-sorbitol buffer - 1x 40% Percoll
Add 1 μl 50 mg/ml TLCK
1 μl 2 mg/ml aprotinin
5 μl 200 mM PMSF
10 μl 10 mg/ml trypsin inhibitor
10 μl 0.5 M EDTA
in 1 ml 40% Percoll - 1x HEPES-sorbitol buffer
Add 1 μl 50 mg/ml TLCK
1 μl 2 mg/ml aprotinin
5 μl 200 mM PMSF
10 μl 10 mg/ml trypsin inhibitor
10 μl 0.5 M EDTA
in 1 ml HEPES-sorbitol buffer - SDS-cOmpleteTM buffer
10 μl 0.5 M EDTA (final 10 mM)
20 μl 25x cOmpleteTM EDTA-free (final 1x)
470 μl SDS-sample buffer - Coomassie Blue stain (store at room temperature)
0.125 g Coomassie Blue R250 (final 0.125% (w/v))
50 ml methanol (final 50% (v/v))
10 ml acetic acid (final 10% (v/v))
ddH2O to 100 ml - Coomassie destain solution (store at room temperature)
45 ml methanol (final 45% (v/v))
10 ml acetic acid (final 10% (v/v))
45 ml ddH2O
Acknowledgments
This protocol was modified from previously published works (Uehara et al., 2016). This work was supported by JSPS KAKENHI Grant Number 17J06506 (to S.U.), 17K07762 (to Y.I.I.) and 15K07843 (to T.I.). The authors declare no conflicts of interest or competing interests.
References
- Inaba, T., Li, M., Alvarez-Huerta, M., Kessler, F. and Schnell, D. J. (2003). atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J Biol Chem 278(40): 38617-38627.
- Jackson, D. T., Froehlich, J. E. and Keegstra, K. (1998). The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J Biol Chem 273(26): 16583-16588.
- Kouranov, A., Chen, X., Fuks, B. and Schnell, D. J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol 143(4): 991-1002.
- Okawa, K., Inoue, H., Adachi, F., Nakayama, K., Ito-Inaba, Y., Schnell, D. J., Uehara, S., and Inaba, T. (2014). Targeting of a polytopic membrane protein to the inner envelope membrane of chloroplasts in vivo involves multiple transmembrane segments. J Exp Bot 65(18): 5257-5265.
- Okawa, K., Nakayama, K., Kakizaki, T., Yamashita, T., and Inaba, T. (2008). Identification and characterization of cor413im proteins as novel components of the chloroplast inner envelope. Plant Cell Environ 31(10): 1470-1483.
- Rolland, V., Badger, M. R. and Price, G. D. (2016). Redirecting the cyanobacterial bicarbonate transporters BicA and SbtA to the chloroplast envelope: Soluble and membrane cargos need different chloroplast targeting signals in plants. Front Plant Sci 7: 185.
- Smith, M. D., Schnell, D. J., Fitzpatrick, L. and Keegstra, K. (2003). In vitro analysis of chloroplast protein import. Curr Protoc Cell Biol Chapter 11: Unit11 16.
- Uehara, S., Adachi, F., Ito-Inaba, Y. and Inaba, T. (2016). Specific and efficient targeting of cyanobacterial bicarbonate transporters to the inner envelope membrane of chloroplasts in Arabidopsis. Front Plant Sci 7: 16.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Uehara, S., Ito-Inaba, Y. and Inaba, T. (2018). Investigating Localization of Chimeric Transporter Proteins within Chloroplasts of Arabidopsis thaliana. Bio-protocol 8(3): e2723. DOI: 10.21769/BioProtoc.2723.
Category
Plant Science > Plant cell biology > Organelle isolation
Plant Science > Plant biochemistry > Protein > Isolation and purification
Cell Biology > Organelle isolation > Chloroplast
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









