- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Immunofluorescence Analysis of Human Endocervical Tissue Explants Infected with Neisseria gonorrhoeae
Published: Vol 8, Iss 3, Feb 5, 2018 DOI: 10.21769/BioProtoc.2720 Views: 7934
Reviewed by: Lokesh KalekarBenoit ChassaingAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantifying Intracellular Distributions of HaloTag-Labeled Proteins With SDS-PAGE and Epifluorescence Microscopy
Julia Shangguan and Ronald S. Rock
Jul 20, 2025 2529 Views

Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1345 Views

An Ex Vivo Lung Histoculture Model for Studying Pulmonary Infection and Immune Response With SARS-CoV-2 as an Example of RNA Virus
Elena V. Maryukhnich [...] Elena J. Vasilieva
Dec 20, 2025 790 Views
Abstract
Colonization and penetration of the epithelium is the infection-initiating route of mucosal pathogens. The epithelium counteracts infection by eliciting host cell responses while maintaining the mucosal barrier function. The obligate human sexually transmitted bacterium Neisseria gonorrhoeae, or gonococcus (GC) infects the female reproductive tract primarily from the endocervical epithelium. Due to lack of an infection model that mimics all aspects of human infections in the female reproductive tract, GC pathogenesis is poorly understood. This protocol takes advantage of the viability and functional integrity of human cervical tissues propagated in culture to generate an ex vivo infection model. This tissue model maintains the nature of the infection target and environment without any manipulation such as immortalization of epithelial cells by viruses. Using immunofluorescence microscopy, the interaction of GC with the endocervical epithelium was analyzed.
Keywords: Neisseria gonorrhoeaeBackground
Neisseria gonorrhoeae (GC) infects human genital epithelium causing gonorrhea, a common sexually transmitted infection. Infections in women can lead to severe complications, such as pelvic inflammatory disease, causing fallopian tube scarring and blockage and predisposition to ectopic pregnancy or infertility. Gonorrhea has reemerged as a critical public health issue due to increased prevalence of antibiotic-resistant strains. Because humans are the only host for GC, a lack of an infection model that mimics all aspects of human infections has been a major obstacle to advance our understanding of GC pathogenesis. We have established a human endocervical tissue explant model and immunofluorescence microscopic analysis to examine the mechanism by which GC infect the human endocervix, the primary site for GC infection in women. This ex vivo model maintains the normal cytoarchitecture and tissue integrity of the endocervical epithelium. Using this model and immunofluorescence analysis, we demonstrate that GC colonizes and penetrates into the endocervical tissue, where they potentially cause symptomatic and disseminated gonococcal infection. GC penetration is enabled by the junction disruption and exfoliation of endocervical epithelial cells in response to GC infection. Taken together, our data show that GC infection in endocervical tissue explants resembles GC infection in vivo observed using patients’ biopsies. In combination with immunofluorescent microscopy, this infection model removes an important roadblock to fully understanding the pathogenesis of GC.
Materials and Reagents
- Cotton swab (Fisher Scientific, catalog number: 23-400-122 )
Manufacturer: Thermo Fisher Scientific, catalog number: 16F0024 . - Carbon steel surgical blades #20 (Aspen Surgical, Bard-Parker, catalog number: 371120 )
- Petri dishes (VWR, catalog number: 25384-302 )
- 6-well tissue culture plates (Corning, Falcon®, catalog number: 353046 )
- Sterile Microcentrifuge tubes (1.7 ml) (Sorenson BioScience, catalog number: 16070 )
- Sterile 15 ml conical tubes (VWR, catalog number: 21008-216 )
- Sterile polyester-tipped applicators (Fisher Scientific, catalog number: 23-400-122 )
- Pipette tips (1,000 μl: VWR, catalog number: 83007-382 ; 200 μl: VWR, catalog number: 53509-007 ; 0.1-10 μl: Fisher Scientific, catalog number: 02-717-133 )
- PAP pen for immunostaining (Sigma-Aldrich, catalog number: Z377821-1EA )
Manufacturer: TED PELLA, catalog number: 22309 . - VWR Micro Slides Superfrost Plus (VWR, catalog number: 48311-703 )
- Zeiss NO.1.5 cover glasses (ZEISS, catalog number: 474030-9000-000 )
- Polysciences flat embedding mold 1.2 x 0.5 x 0.4 cm (Length x Width x Depth) (Polysciences, catalog number: 02615 )
- MS11 Neisseria gonorrhoeae strain(s) (kindly provided by Dr. Herman Schneider, Walter Reed Army Institute for Research)
- Hank’s balanced salt solution (HBSS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14025092 )
- Liquid nitrogen (Roberts Oxygen Company)
- Hoechst 33342, trihydrochloride, trihydrate, 10 mg/ml (Thermo Fisher Scientific, InvitrogenTM, catalog number: H3570 )
- Alexa Fluor 488 goat anti-mouse IgG1 (Thermo Fisher Scientific, Invitrogen, catalog number: A-21121 )
- Mouse anti-human ZO1 (BD, Transduction LaboratoriesTM, catalog number: 610966 )
- Goat anti-GC antibody (Lab generated)
- DyLight 633 Antibody Labeling Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 53046 )
- Aqua Poly/Mount (Polysciences, catalog number: 18606 )
- Nail polish (Electron Microscopy Science, catalog number: 72180 )
- O.C.T. Compound (SAKURA, Tissue-Tek, catalog number: 4583 )
- Penicillin G sodium salt (Sigma-Aldrich, catalog number: P3032-25MU )
- Streptomycin sulfate (Sigma-Aldrich, catalog number: S9137 )
- Leibovitz’s (1x) L-15 medium (Thermo Fisher Scientific, GibcoTM, catalog number: 21083027 )
- CMRL medium 1066 (1x) (Thermo Fisher Scientific, GibcoTM, catalog number: 11530037 )
- Hydrocortisone 21-hemisuccinate sodium (Sigma-Aldrich, catalog number: H2270-100MG )
- Bovine insulin (Akron Biotech, catalog number: AK8213-0100 )
- L-glutamine 200 mM (100x) (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10082147 )
- DifcoTM GC medium base (BD, DifcoTM, catalog number: 228950 )
- Agar (United States Biological, catalog number: A0930 )
- L-glutamine, White Crystals or Crystalline Powder (Fisher Scientific, catalog number: BP379-100 )
- Ferric nitrate, nonahydrate (Sigma-Aldrich, catalog number: 254223-10G )
- Thiamine pyrophosphate (Sigma-Aldrich, catalog number: C8754-5G )
- Glucose, as Dextrose anhydrous (Fisher Scientific, catalog number: BP350-1 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S671-10 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333-500G )
- Sodium phosphate dibasic (Na2HPO4) (Fisher Scientific, catalog number: S374-1 )
- Potassium phosphate monobasic (KH2PO4) (Fisher Scientific, catalog number: BP329-1 )
- Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A144-212 )
Manufacturer: Thermo Fisher Scientific, catalog number: FLA144212 . - Paraformaldehyde 16% solution (Electron Microscopy Science, catalog number: 15710 )
- Sucrose (Merck, catalog number: SX1075-1 )
- Gelatin from porcine skin (Sigma-Aldrich, catalog number: G1890-500G )
- Gelatin from cold water fish skin (FSG) (Sigma-Aldrich, catalog number: G7765-250ML )
- Triton X-100 (Sigma-Aldrich, catalog number: X100-100ML )
- 100x penicillin/streptomycin stock (see Recipes)
- Cervical tissue culture medium (see Recipes)
- GCK agar plate (see Recipes)
- 100x Kellogg’s supplement (see Recipes)
- 1x phosphate buffer solution (PBS) (see Recipes)
- 4% paraformaldehyde (PFA) (see Recipes)
- 5% and 15% sucrose solution (see Recipes)
- 7.5% and 20% gelatin solution (see Recipes)
- Staining solution (see Recipes)
Equipment
- Biosafety cabinet (NU-425-600 Class II, A2 Laminar Flow Biohazard Hood) (Nuaire, model: NU-425-600 Class II, A2 , catalog number: 32776)
- Spectrophotometer Ultrospec 2000 UV (Pharmacia Biotech, model number: 80-2106-00 )
- Heating block (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SP18425Q )
- Stainless steel forceps (VWR, Dissecting forceps)
- Multi-Purpose rotator (Multi-Purpose Rotator) (Barnstead International Lab-line, Thermo Fisher Scientific, Thermo ScientificTM, model: Model 2309 )
- CO2 incubator (Fisher Scientific, model: Isotemp Incubator Model 3530 )
- Confocal microscope (Carl Zeiss, model: LSM 710 )
- Cryostat (Thermo Fisher Scientific, model: Microm HM 550-388114 )
- Slide holder (Newcomer Supply, catalog number: 6841 )
Software
- NIH ImageJ and ZEN software
- GraphPad Software (La Jolla, CA)
Procedure
- Sample collection and shipping
Cervical tissues (~5 g/piece) from patients undergoing voluntary hysterectomy are obtained in sterile DMEM with 1x penicillin/streptomycin (see Recipes) at 4 °C within 24 h post-surgery (Merbah et al., 2011), coordinated by National Disease Research Interchange (NDRI) (an institutional review board must approve the research protocol involving human tissues. Please verify with your own institutional review board for the regulation). - Tissue preparation
The tissue explants were cultured based on a method described by Schürch et al. (1978).- Rinse tissues three times with cold HBSS.
- Rinse tissues three times with cold Leibovitz’s (1x) L-15 medium.
- Remove mucus gently using a cotton swab.
- Remove muscle tissues using a carbon steel surgical blade (Figure 1A, black dashed line).
- Separate the endocervical region from the rest of the cervix tissue explant using a carbon steel surgical blade (Figure 1A). Store the endocervix for the next steps of the protocol while the rest of the cervix is discarded.
- Rinse endocervical tissue explants three times with cervical tissue culture medium.
- Incubate endocervical tissue explants in a 6-well tissue culture plate, one piece per well, in cervical tissue culture medium at 37 °C with 5% CO2 for 24 h.
- Take a picture of tissue along with its well.
- Determine the area of the tissue luminal surface (dotted line in Figure 1B) by measuring the tissue to well size ratio using NIH ImageJ software.
- Wash the endocervical tissue explants three times with antibiotic-free cervical tissue culture medium (see Recipes), and incubate the tissue in the antibiotic-free media at 37 °C with 5% CO2 for 24 h before bacterial inoculation.
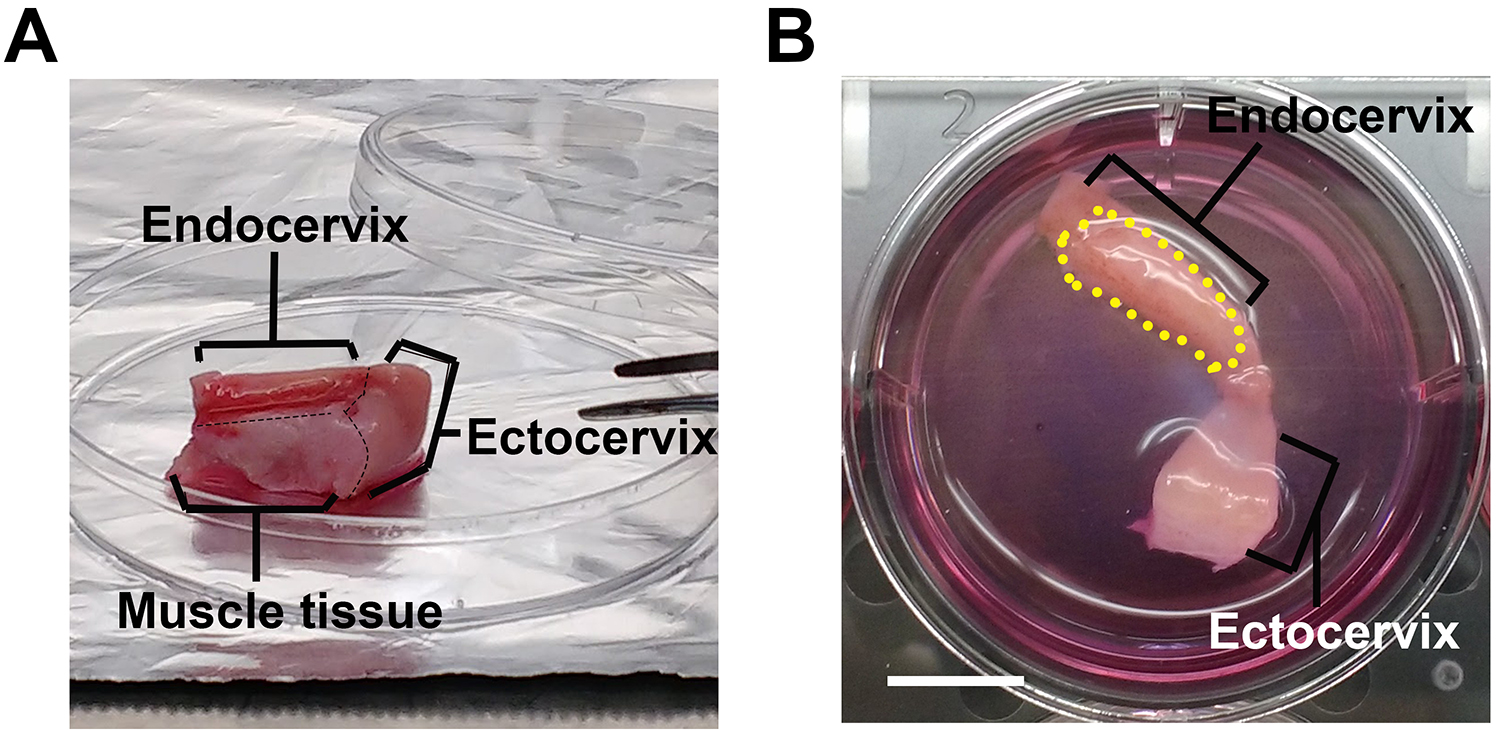
Figure 1. Endocervical tissue explant in culture. The endocervical tissues (A), after removing mucus, muscle tissue and other parts of the cervix, are trimmed into the desired size (B) and cultured in the cervical tissue culture medium in a six-well plate. The black dashed lines indicate the cutting line on tissues and the yellow dash line circles the luminal surface of the endocervix. Scale bar = 10 mm.
- Rinse tissues three times with cold HBSS.
- Infection of the endocervix with GC
- Grow GC on GCK agar plate (see Recipes) for 16-18 h.
- Collect GC by using a sterile applicator to swab GC from the plate and re-suspend GC in pre-warmed antibiotic-free cervical tissue culture media. Determine the number of bacteria by spectrophotometry at a wavelength of 650 nm (an OD650 of 1 = 1 x 109 CFU/ml).
- Estimate the number of epithelial cells by dividing the luminal surface area of a tissue explant by the average luminal surface area of individual endocervical epithelial cells (25 µm2) (Figure 3D).
- Add an aliquot of the GC suspension (1 ml) directly onto each tissue piece to make an MOI = 10 bacteria to 1 epithelial cell and incubated at 37 °C with 5% CO2 for 3 min.
- Add 1 ml of antibiotic-free cervical tissue culture medium (see Recipes) to each of the above tissue pieces.
- Incubate tissue explants at 37 °C with 5% CO2 with gentle shaking for 24 h. Rinse infected tissue explants 6 times with 3 ml antibiotic-free cervical tissue culture medium at 6 and 12 h to remove un-adhered bacteria and replace with 2 ml of fresh media without bacteria.
- Grow GC on GCK agar plate (see Recipes) for 16-18 h.
- Tissue preparation for cryosection–Fixation
- Rinse tissue explants three times with 3 ml PBS (see Recipes).
- Fix tissue explants with 3 ml 4% paraformaldehyde (in PBS) (see Recipes) for 1 h at room temperature.
- Rinse tissue explant three times with PBS.
- Cut tissues into a desired size, approximately 1.0 x 0.5 x 0.2 cm (Length x Width x Depth), using a razor blade.
- Rinse tissue explants three times with 3 ml PBS (see Recipes).
- Tissue preparation for cryosection–Infiltration & Embedding
- Incubate tissue explants in 5 ml of 5% sucrose in PBS (see Recipes) at room temperature for 30 min until the explants settle to the bottom of the tube.
- Carefully remove the 5% sucrose and resuspend the tissue in 5 ml of 15% sucrose (in PBS) (see Recipes) and incubated overnight at 4 °C.
- Carefully remove the sucrose and resuspend tissue explants in 7.5% porcine skin gelatin in PBS (see Recipes) for 4 h at 37 °C.
- Carefully remove the gelatin solution and resuspend tissue explants in 5 ml pre-warmed 20% porcine skin gelatin in PBS (see Recipes) and incubated for 2 h at 37 °C.
- Pre-warm a flat embedding mold (Figure 2A) for each piece of tissues on a heating block at 37 °C.
- Fill the bottom 1/3 of molds with 20% gelatin and let it solidify at room temperature.
- Put one tissue piece per mold and top with 20% gelatin at 37 °C. Arrange the orientation of the tissue using pipette tips as shown in Figure 2A to have luminal surface at the top. Make sure the tissue piece is fully embedded in gelatin (Figure 2B).
- Let samples sit for 5 min at room temperature and then on ice block for 10 min.
- Use long forceps to place the mold containing tissues into liquid nitrogen vapor until the gelatin turns white.
- Dip the mold into liquid nitrogen for a few seconds till the samples are all frozen.
- Push sample blocks out of molds by warming the bottom of the mold at 42 °C on a heating block for 30 sec.
- Store the embedded samples in conical tubes at -80 °C.
- Incubate tissue explants in 5 ml of 5% sucrose in PBS (see Recipes) at room temperature for 30 min until the explants settle to the bottom of the tube.
- Cryosection
- Place a sample block on the stage of a cryostat in an orientation, which enables sectioning through both the luminal surface and the subepithelium (Figure 2C).
- Section sample blocks in 40 μm thickness until reaching the tissue.
- Switch to desired section thickness ranging from 10-40 μm.
- Collect sample sections by attaching them to glass slides (Figure 2C).
- Store cryosections on glass slides at 4 °C for the same day processing or -20 or -80 °C for future processing.
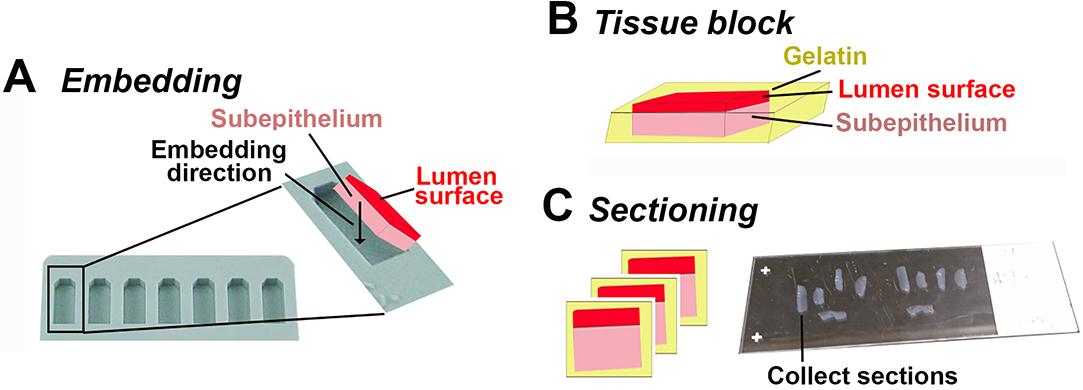
Figure 2. Embedding and sectioning of endocervical tissues. A-B. Endocervical tissues are embedded with the luminal side up in 20% gelatin. Black arrow, the direction that the tissue is positioned. C. Endocervical tissue blocks are sectioned transversally through both the luminal surface and the subepithelial tissue. Endocervical tissue sections are collected using super frost plus slides for immunofluorescence staining.
- Place a sample block on the stage of a cryostat in an orientation, which enables sectioning through both the luminal surface and the subepithelium (Figure 2C).
- Immunofluorescence staining for imaging
- Incubate tissue sections on glass slides in a slide holder with PBS at 42 °C for 10 min to remove gelatin.
- Wash tissue sections three times with PBS.
- Aspirate excess PBS and draw an aqueous barrier circle round tissue sections using a pap pen.
- Fill the circle with the staining solution (see Recipes) and incubate at room temperature for 1 h for permeabilizing and blocking.
- Incubate tissue sections with a primary antibody in the staining solution at room temperature for 1 h. (Table 1)
Table 1. The antibodies/chemicals and the final concentration used in Figures 2 and 3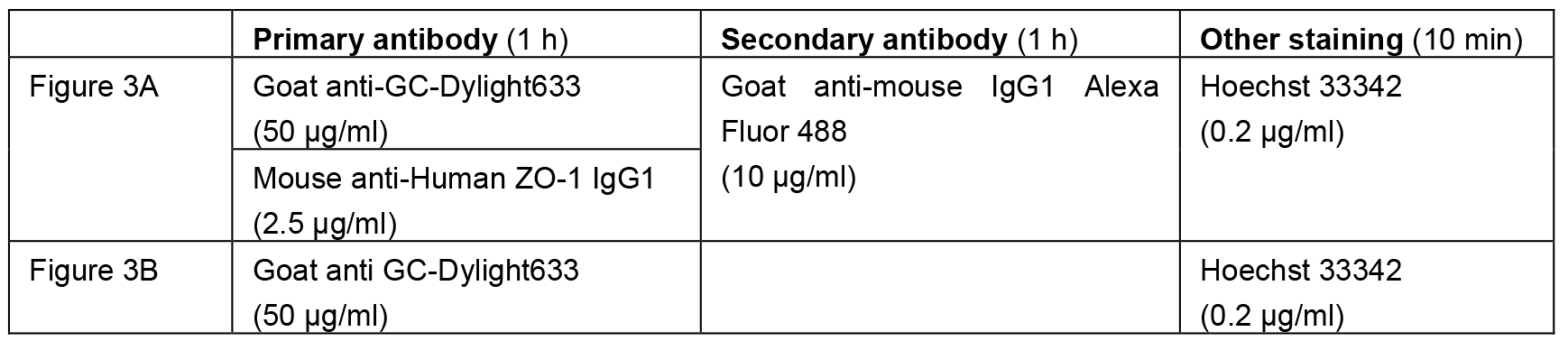
Note: Fluorescent phalloidin and immunofluorescence E-cadherin staining can also be included to assess the cellular location of GC. - Rinse tissue sections three times with the staining solution.
- Incubate tissue sections with a secondary antibody in the staining solution at room temperature for 1 h. (Table 1)
- Rinse tissue sections three times with the staining solution.
- Incubate tissue sections with Hoechst 33342 (0.2 μg/ml) in the staining solution at room temperature for 10 min. (Table 1)
- Rinse tissue sections three times with the staining solution.
- Mount tissue sections using Aqua/poly mounting media and No.1.5 or No. 1 coverslip glass. Dry for 5 min before sealing the edge of coverslip using nail polish.
- Slides are now ready for imaging or can be stored at 4 °C for up to 2 months.
- Incubate tissue sections on glass slides in a slide holder with PBS at 42 °C for 10 min to remove gelatin.
- Imaging
- Acquire images using a ZEISS LSM 710 confocal microscope (an equivalent microscope can be used).
- Acquire Z-stack of 0.37 μm slices from the top to the bottom of a tissue section (Figure 3A).
- Reconstitute Z-stack images into 3D-images using ImageJ or ZEN software (Figure 3B), which allows obtaining both the transversal (Figure 3C) and luminal views (Figure 3D) of the endocervical epithelium.
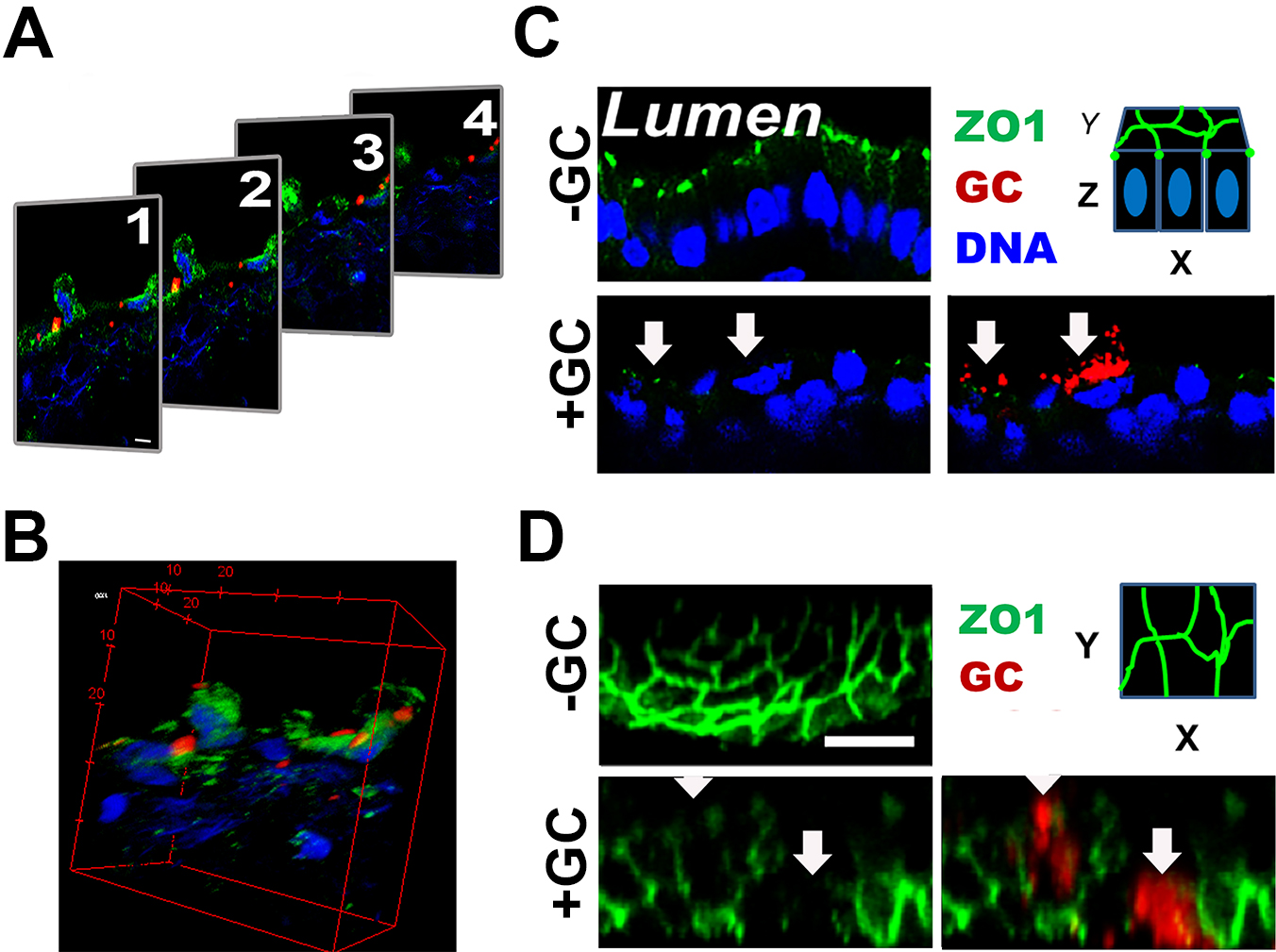
Figure 3. 3D-reconstruction of fluorescence images. Cryo-sections of endocervical tissue explants were stained for ZO-1 (green), GC (red), and DNA (blue). Z-stack images were taken using a confocal fluorescent microscope. Shown are series of Z-stack images (A) and their 3D reconstruction (B). Images of XZ view (C) and XY view (D) of the luminal surface are then generated for publication. The red dashed line circles the luminal surface of an epithelial cell (~25 µm2). Scale bar = 10 µm.
- Acquire images using a ZEISS LSM 710 confocal microscope (an equivalent microscope can be used).
Data analysis
In our published studies, we quantitatively analyzed the penetration of GC into the subepithelium of the endocervix and the exfoliation of endocervical epithelial cells.
- To estimate the level of GC penetration into the subepithelium, the number of non-exfoliated endocervical epithelial cells (in visible epithelial monolayers) with GC staining at the basal side and the total number of GC-associated epithelial cells in randomly acquired image are visually counted to calculate the percentage of infected epithelial cells with GC penetration into the epithelium (Figure 4A).
- To quantify epithelial exfoliation, the number of endocervical epithelial cells localized on the top of the monolayer (exfoliated) and the total number of endocervical epithelial cells were counted by visual inspection in each randomly acquired image to determine the percentage of exfoliated cells (Figure 4B).
- Quantified values are plotted into column graphs, and statistical significance is assessed using the Student’s t-test and one-way ANOVA by Prism (GraphPad Software, La Jolla, CA).
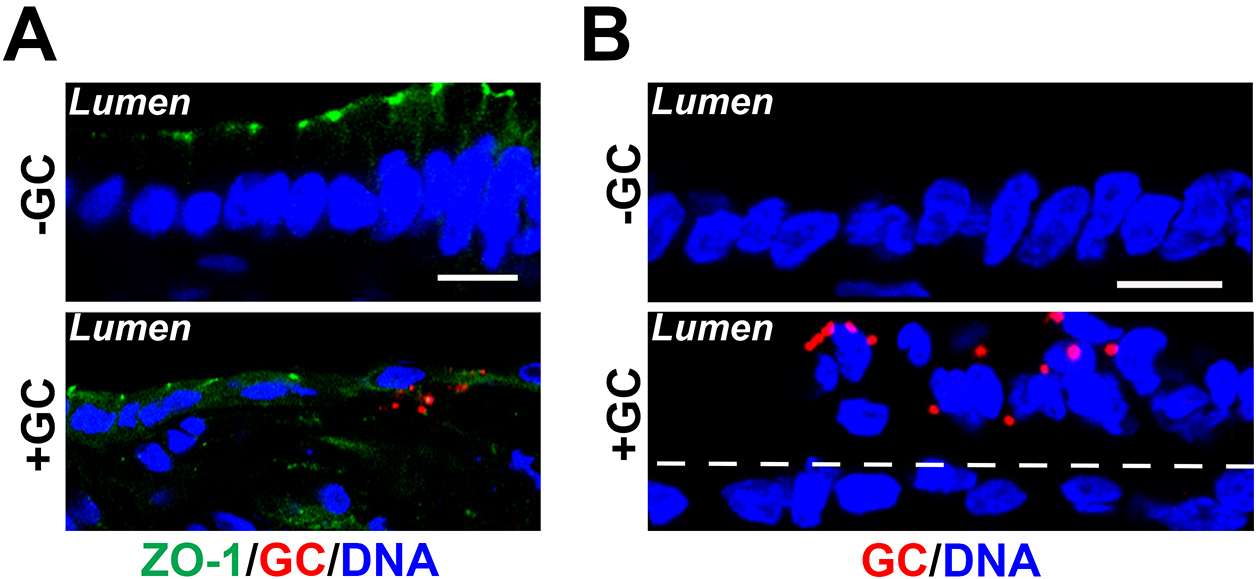
Figure 4. Fluorescent image analysis of GC infection in the endocervical tissue explant. Cryo-sections of cervical tissue explants were stained for ZO-1 (green), GC (red), and DNA (blue). Shown are representative images that cross both the luminal and basal surfaces of epithelial cells (Scale bars = 10 μm). A. GC subepithelial penetration was quantified using immunofluorescence images as the percentage of epithelial cells with basal GC staining (arrows) among the total number of GC-associated epithelial cells. B. The average percentage of exfoliated epithelial cells was determined by the number of epithelial cells localizing above the endocervical epithelium (white dash lines) versus the total number of epithelial cells, based on the nuclear staining.
Recipes
- 100x penicillin/streptomycin (P/S) stock
10,000 UI/ml penicillin
10,000 μg/ml streptomycin
Phenol Red-free DMEM/F12 medium
Store at -20 °C - Cervical tissue culture medium
CMRL-1066 medium
0.1 μg/ml hydrocortisone 21-hemisuccinate sodium
1 μg/ml bovine insulin
2 mM L-glutamine
5% FBS
1x penicillin-streptomycin (P/S)
Store at 4 °C - GCK agar plate (for 1 L)
35 g Difco GC medium base
6 g agar
10 ml 100x Kellogg’s supplement
Store at 4 °C - 100x Kellogg’s supplement (for 1 L)
5 g L-glutamine Crystalline Powder
0.5 g ferric nitrate, anhydrous
0.02 g thiamine pyrophosphate
400 g glucose
Store at 4 °C - 1x phosphate buffer solution (PBS) (for 1 L)
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4
0.24 g KH2PO4
Adjust pH to 7.4 with HCl
Store at 4 °C - 4% paraformaldehyde (PFA)
30 ml 16% paraformaldehyde stock
10 ml 1x PBS
Store at -20 °C - 5% and 15% sucrose solution
5 g (for 5%) or 15 g (for 15%) sucrose
100 ml 1x PBS
Store at -20 °C - 7.5% and 20% gelatin solution
7.5 g (for 7.5%) or 20 g (for 20%) porcine skin gelatin
100 ml 1x PBS
Heat to solubilize
Store at -20 °C - Staining solution
1x PBS
0.66% Fish skin gelatin (FSG)
0.2% Triton X-100
Store at 4 °C
Acknowledgments
We would like to thank NDRI for tissue collection. This work was supported by NIH grants (RO1 AI068888, R21 AI103797, and RO1 AI123340 to WS and DCS). This protocol was partially adapted from previously published work by Merbah et al. (2011) and Schürch et al. (1978). The authors declare no conflicts of interest within this work.
References
- Merbah, M., Introini, A., Fitzgerald, W., Grivel, J. C., Lisco, A., Vanpouille, C. and Margolis, L. (2011). Cervico-vaginal tissue ex vivo as a model to study early events in HIV-1 infection. Am J Reprod Immunol 65(3): 268-278.
- Schürch, W., McDowell, E. M. and Trump, B. F. (1978). Long-term organ culture of human uterine endocervix. Cancer Res 38(11 Pt 1): 3723-3733.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, L., Yu, Q., Stein, D. C. and Song, W. (2018). Immunofluorescence Analysis of Human Endocervical Tissue Explants Infected with Neisseria gonorrhoeae. Bio-protocol 8(3): e2720. DOI: 10.21769/BioProtoc.2720.
Category
Microbiology > Microbe-host interactions > Ex vivo model
Immunology > Mucosal immunology > Epithelium
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









