- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Neisseria meningitidis Adherence to Human Epithelial Cells by Colony Counting
(*contributed equally to this work) Published: Vol 8, Iss 3, Feb 5, 2018 DOI: 10.21769/BioProtoc.2709 Views: 8749
Reviewed by: Chao JiangAndrea PuharAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

HS–GC–MS Method for the Diagnosis of IBD Dynamics in a Model of DSS-Induced Colitis
Olga Yu. Shagaleeva [...] Natalya B. Zakharzhevskaya
Mar 20, 2025 3108 Views

In Vitro Co-culture of Bacterial and Mammalian Cells to Investigate Effects of Potential Probiotics on Intestinal Barrier Function
Ajitpal Purba [...] Dulantha Ulluwishewa
Jun 20, 2025 2515 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views
Abstract
To cause an infection, the human specific pathogen Neisseria meningitides must first colonize the nasopharynx. Upon tight interaction with the mucosal epithelium, N. meningitidis may cross the epithelial cellular barrier, reach the bloodstream and cause sepsis and/or meningitis. Since N. meningitidis niche is restricted to humans the availability of relevant animal models to study host-pathogen interactions are limiting. Therefore, most findings that involve N. meningitidis colonization derive from studies using cultured human cell lines. Human epithelial cells have been successfully used to examine and identify molecular effectors involved in initial adherence of the pathogen. Here, we describe a standard protocol to quantify the adherence of N. meningitidis to epithelial pharyngeal FaDu cells. Colony counts of cell lysates collected after infection are used to quantify adherence to the epithelial cells.
Keywords: NeisseriaBackground
Upon entry to a new host, adherence to specific host tissues serves as an important step in bacterial pathogenesis. Molecular interaction between bacterial adhesins and receptors on the host cell surface determines colonization sites (Soto and Hultgren, 1999). The epithelial layer in the nasopharynx forms the first cellular barrier that the human restricted pathogen N. meningitidis encounters and colonizes asymptomatically. Tight adherence and interaction with the host cells can lead to penetration of the epithelium and entry into the bloodstream, resulting in life-threatening sepsis and/or meningitis (Stephens, 2009). Long filaments extending from the bacterial membrane, called type IV pili (Tfp), containing PilC1 tip-located adhesin play a key role in initial adherence of N. meningitidis to the nasopharyngeal epithelium (Marceau et al., 1995; Rudel et al., 1995). Tfp does not only promote interaction with host cells but is also involved in the development of bacterial aggregates, that can contribute to a high level of adherence and resistance against shear stress (Helaine et al., 2005; Mikaty et al., 2009; Engman et al., 2016). Apart from the Tfp, other surface expressed molecules like the opacity proteins, LPS, NadA, NhhA, App and MspA have been shown to affect the level of adhesion to the epithelial surface (Hill et al., 2010).
Animal models to study N. meningitidis colonization are limiting due to human host specificity. Consequently, the majority of the studies over the years have relied on cultured human cell lines (Merz and So, 2000). Here, we provide a step-by-step protocol adapted from Sigurlásdóttir et al. (2017) to quantify adherence of N. meningitidis to human epithelial pharyngeal FaDu cells in culture. In the following protocol, human epithelial cells are infected with both wild-type and an adhesion-deficient ΔpilC1 strain at a multiplicity of infection (MOI) of 10 for an incubation time of 4 h. The procedure described herein for N. meningitidis adherence to cultured epithelial cells could be easily applicable to a range of different bacterial species and cell lines with adaptation of the growth media (de Klerk et al., 2017).
Materials and Reagents
- Lab coat and protective gloves
- Marker pen
- Cell culture flasks T75 (SARSTEDT, catalog number: 83.1813.001 )
- Cell culture plates 24-well (SARSTEDT, catalog number: 83.1836 )
- Serological pipettes
2 ml pipette (SARSTEDT, catalog number: 86.1252.001 )
5 ml pipette (SARSTEDT, catalog number: 86.1253.001 )
10 ml pipette (SARSTEDT, catalog number: 86.1254.001 )
25 ml pipette (SARSTEDT, catalog number: 86.1685.001 )
- Sterile plastic loops
1 µl plastic loops (SARSTEDT, catalog number: 86.1567.050 )
10 µl plastic loops (SARSTEDT, catalog number: 86.1562.050 )
- Falcon tubes
15 ml tubes (SARSTEDT, catalog number: 62.554.502 )
50 ml tubes (SARSTEDT, catalog number: 62.547.254 )
- Pipette tips
20-200 µl capacity (SARSTEDT, catalog number: 70.760.502 )
50-1,000 µl capacity (SARSTEDT, catalog number: 70.762.100 )
- 5 µm pore filter (VWR, catalog number: 514-4106 )
- Cell culture plates 96-well (SARSTEDT, catalog number: 83.3924 )
- Bacteriological Petri plates, 92 x 16 mm (SARSTEDT, catalog number: 82.1473 )
- 5 ml syringe (VWR, catalog number: 613-3940 )
- 250 ml vacuum filtration unit, 0.22 μm (SARSTEDT, catalog number: 83.1822.001 )
- Bacterial strain: Neisseria meningitidis serogroup C strain FAM20 wild-type and ΔpilC1 (Rahman et al., 1997). The bacterial strain FAM20 is a nalidixic acid-resistant mutant of FAM18 that is available at ATCC (ATCC, catalog number: 700532 )
Note: The bacterial stocks are stored in 25% glycerol:75% GC liquid medium (see Recipes) at -80 °C.
- Cell line: pharyngeal epithelial cell line FaDu (ATCC, catalog number: HTB-43 )
Note: The cell line is stored in 90% FBS:10% DMSO at -140 °C.
- 70% ethanol
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- DMEM high glucose, GlutaMAXTM Supplement, pyruvate (Thermo Fisher Scientific, catalog number: 31966047 )
- Fetal bovine serum (FBS), heat inactivated (Sigma-Aldrich, catalog number: F9665 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418 )
- Phosphate-buffered saline (PBS), 10x concentrated (Statens Veterinärmedicinska Anstalt, catalog number: 992442 )
- GC agar medium base (NEOGEN, Acumedia, catalog number: 7104A )
- D-glucose (Sigma-Aldrich, catalog number: G8270 )
- L-glutamine (Sigma-Aldrich, catalog number: G8540 )
- Ferric nitrate (Sigma-Aldrich, catalog number: F3002 )
Note: This product has been discontinued.
- Cocarboxylase (Sigma-Aldrich, catalog number: C8754 )
- Saponin (Sigma-Aldrich, catalog number: S7900 )
- Protease peptone (Oxoid, catalog number: LP0085 )
- Starch, soluble (Sigma-Aldrich, catalog number: S9765 )
- Potassium phosphate dibasic (Sigma-Aldrich, catalog number: 60353 )
- Potassium phosphate monobasic (Sigma-Aldrich, catalog number: 60218 )
- Sodium chloride (Sigma-Aldrich, catalog number: S3014 )
- Trypsin-EDTA (0.5%), no phenol red, 10x (Thermo Fisher Scientific, GibcoTM, catalog number: 15400054 )
- GC agar plates (see Recipes)
- Kellogg’s supplement (see Recipes)
- Cocarboxylase solution (see Recipes)
- 2x trypsin (see Recipes)
- 1% saponin (see Recipes)
- Phosphate-based GC liquid medium (see Recipes)
Equipment
- Class II biosafety cabinet (e.g., Esco Micro, model: Airstream® Max )
- Incubator at 37 °C and with a 5% CO2 environment (e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: HeracellTM 150i )
- Cell culture hood (e.g., ESCO laminar flow cabinet, Esco Micro, model: Airstream® Gen 3 )
- Inverted microscope (e.g., Carl Zeiss, model: Axiovert 40 C )
- Spectrophotometer (e.g., Bio-Rad Laboratories, model: SmartSpec Plus )
- Hemocytometer (e.g., VWR, catalog number: 631-0923 )
- Pipette boy (e.g., Fisher Scientific, model: Fisherbrand Electric Pipet Controller )
- Pipettes
10-100 µl capacity (e.g., Eppendorf, catalog number: 4924000053 )
100-1,000 µl capacity (e.g., Eppendorf, catalog number: 4924000088 )
- Multichannel pipette, 10-100 µl capacity (e.g., Eppendorf, catalog number: 3125000036 )
- Water bath set to 37 °C (e.g., Grant Instruments, model: Sub Aqua Pro, catalog number: SAP12 )
- Centrifuge
- 1 L flask
- Autoclave
Procedure
Caution: N. meningitidis is designated a class II pathogen. Preparation of all bacterial cultures should be performed carefully in a Class II biosafety cabinet and according to national biosafety guidelines. Wear a lab coat and protective gloves during handling of the bacteria. Decontaminate work surface with 70% ethanol before and after use. Wash hands after removing gloves. All material that has been in contact with bacteria should be discarded as an Infectious/Biohazardous Waste.
Day 1
- Growth of N. meningitidis
- Wild-type and ΔpilC1 mutant bacteria are streaked from glycerol stocks on GC agar plates (see Recipes).
- Incubate plates overnight for 16-18 h at 37 °C in an incubator with a 5% CO2 environment.
- Wild-type and ΔpilC1 mutant bacteria are streaked from glycerol stocks on GC agar plates (see Recipes).
- Preparation of cell cultures (work in a cell hood under sterile conditions)
Note: The cell line, FaDu, is maintained in DMEM containing 10% FBS and split at ratio 1:3 or 1:6 every 3-5 days. The cells from a fully confluent cell flask of 75 cm2 (T75) are required to prepare one 24-well plate the day before the experiment. All volumes indicated below in Steps 2a-2d are given for one 75 cm2 flask as starting material. The cell line is categorized biosafety level 1. - Prewarm DMEM supplemented with 10% FBS, sterile 1x PBS and 2x trypsin (see Recipes) for at least 30 min in a 37 °C water bath.
- Wash the cells once with 5 ml of 1x PBS.
- Add 1 ml of 2x trypsin and allow the cells to detach in a 37 °C incubator for 5 min. Examine the cells under an inverted microscope and make sure that no aggregates are visible.
- Resuspend the detached cells in 20 ml of pre-warmed DMEM supplemented with 10% FBS. Use a 10 ml serological pipette to mix the solution until homogenized.
- Count number of cells per ml with a hemocytometer. Seed cells at a density of 1.5 x 105 cells/well to a 24-well plate.
- Incubate the cell culture plate overnight and allow to adhere and grow to 100% confluence (2 x 105) at 37 °C in an incubator with a 5% CO2 environment. It is important to examine cell confluence and check for contamination before using cells in an adhesion assay.
Note: Having the cultured epithelial cells at 100% confluence, eliminates the possibility of N. meningitidis being stuck in between the intracellular gap. The procedure ensures that the adherence observed is solely due to N. meningitidis adhered to the epithelial cells and not to any abiotic surface.
Day 2
- Adhesion assay (work in a Class II biosafety cabinet)
Note: Before the adhesion assay, the OD600 nm that is equal to the viable count 108 cfu/ml has to be determined for the bacterial stock used. This can be determined by serial dilutions and plating to GC plates. For the bacterial stocks used in this experiment, OD600 nm: 0.36 is equal to 108 cfu/ml.- Prewarm DMEM. Before the experiment wash the cells gently once and add 800 µl of fresh DMEM. While preparing bacterial cultures, keep the cell culture plate in an incubator at 37 °C with a 5% CO2 environment.
Note: During the adhesion assay, DMEM is used without FBS supplementation.
- For each bacterial strain, pick bacteria (half fill a 10 μl loop) from the GC agar plate grown overnight and resuspend in a 15 ml Falcon tube containing 2 ml of DMEM. Use 1 ml pipette to pipette up and down 4 times to mix the bacterial solutions. Let the solution stand at room temperature for 3 min to let the largest bacterial aggregates sediment to the bottom of the tube.
Note: The N. meningitidis bacterial cultures that are used in adhesion assays should never be vortexed as this might shear the Tfp and influence the results.
- Take 1 ml of the supernatant and transfer to a 50 ml Falcon tube containing 5 ml DMEM. Filter the bacterial solutions through 5 µm pore filter.
Note: The level of N. meningitidis aggregation can influence the number of adhered bacteria. Therefore we chose to filter the bacterial suspension, to have only single bacterial cells in the solution before starting the adhesion assay. Filtering can thus prevent artefactual variations in the level of adhesion that would stem from differences in re-suspension of the bacteria. (Eriksson et al., 2012; Sigurlásdóttir et al., 2017).
- Measure the absorbance of the bacterial solution with a spectrophotometer. Adjust the absorbance to OD600 = 0.36 (equivalent to 108 cfu/ml) by diluting the bacterial solution with DMEM.
- After the bacterial solution has been adjusted to OD600 = 0.36, dilute the solution 10-fold (900 µl:100 µl, equivalent 107 cfu/ml). Pipette the solution up and down 4 times with a 1 ml pipette. Infect cells at MOI of 10 by adding 200 µl of the bacterial solution (2 x 106 cfu/ml) to relevant wells of the cell culture plate. The total volume in the wells should be 1 ml since 800 µl of fresh DMEM is added as indicated in Step 1a. To ensure equal distribution of the inoculum, add 200 µl of the bacterial solutions gently drop by drop apart from each other within the well. Each bacterial strain should be added preferably in triplicates, at least in duplicates.
Note: The multiwell plate containing infected cells can be centrifugated (200 x g for 5 min) before infection starts. Centrifugation can enhance contact between host and bacteria. If adhesion assay is performed in parallel to time-lapse imaging, then centrifugation can be useful to accelerate the binding of bacteria to host cells (Eriksson et al., 2012; Sigurlásdóttir et al., 2017).
- Transfer the cell culture plate to a 37 °C incubator with a 5% CO2 environment. Incubate for 4 h.
- To ensure the viable count for every experiment performed, plate serial dilutions of the prepared bacterial cultures for each strain in duplicates immediately after the incubation of the adhesion assay starts. Plate to GC plates and incubate overnight in a 37 °C incubator with 5% CO2 overnight. The actual MOI for every experiment should be calculated from the viable count.
- During the 4 h incubation, leave 2 GC agar plates in the biosafety hood turned upside down to dry. Divide the GC agar plates into 12 squares with the help of a marker pen (Figure 1A). The division will help to spot 12 different dilutions on the same plate, thereby saving media (Figure 1B). As an alternative approach that will require 24 GC plates, spread 100 µl of each dilution to a GC agar plate by using a 10 µl loop. Prepare for serial dilutions, prepare five 1:10 dilution series in DMEM with a total volume of 200 µl (180 µl DMEM) in a 96-well plate (Figure 2).
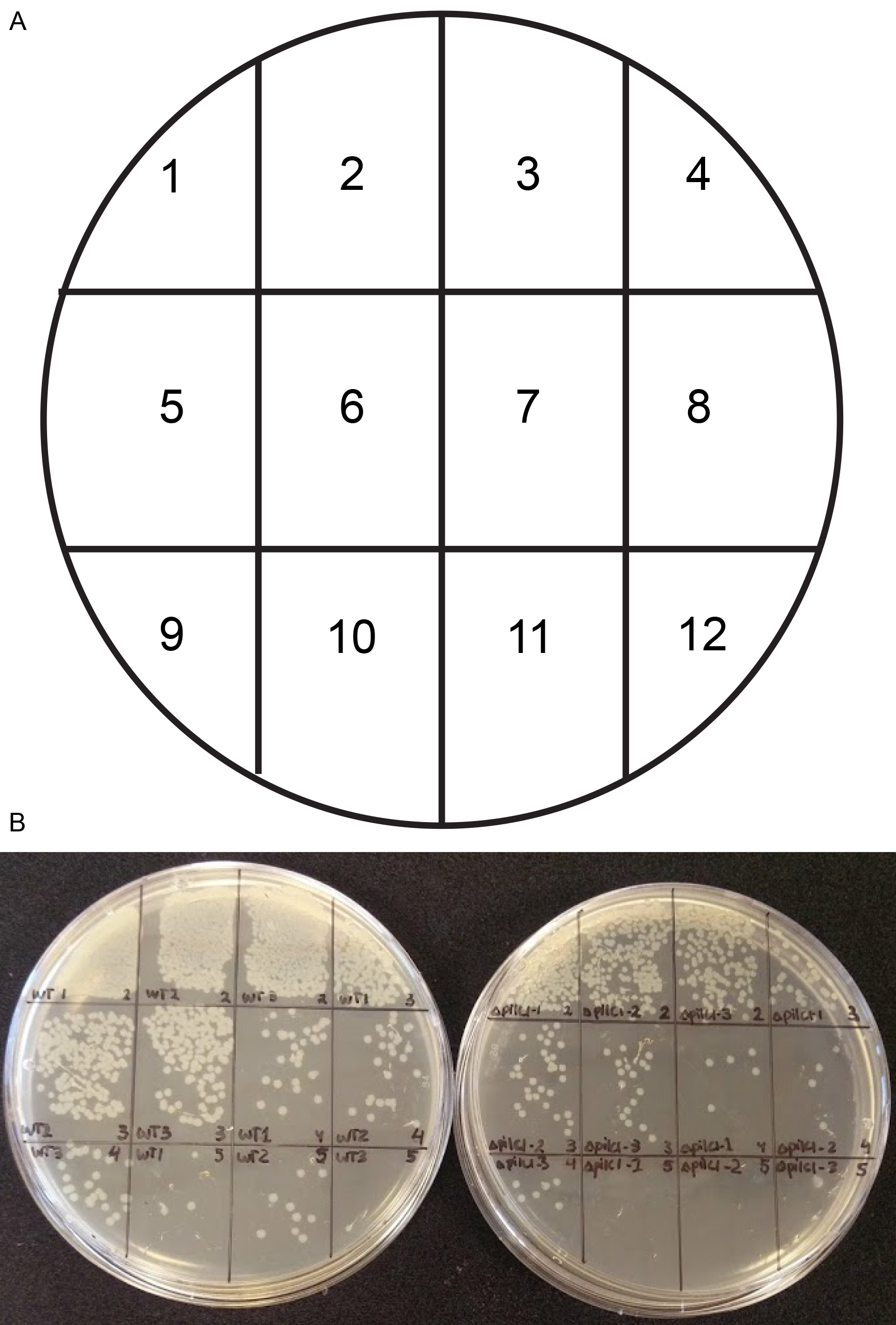
Figure 1. Template for plating 12 samples on a GC agar plate. A. Illustration of the GC agar plate divided into 12 squares. B. For each bacterial strain (WT–wild type and ΔpilC1 in triplicates), four (-2 to -5) dilutions per sample are divided into the 12 squares on the agar plate.
- After the 4 h incubation, take the cell culture plate out from the incubator. Work in a Class II biosafety cabinet. Gently wash unbound bacteria away three times by removing all cell culture media and adding 1 ml fresh DMEM to the wells. Carefully try not to detach the cells. No unbound bacteria should be visible under the microscope when washing is finished. Under the microscope, unbound bacteria would appear as free bacteria floating in the growth media.
- Lyse cells with 1 ml of 1% saponin (see Recipes) in DMEM for 10 min in a 37 °C incubator.
- Take the plate out of the incubator and check lysis of the cells through a microscope. If cells have not detached completely from the bottom of the plate, use the end of a sterile plate loop to scrape the cells from the bottom. Use a 1 ml pipette and mix the cell lysates up and down 4 times and transfer 200 µl to row A of the 96-well plate (Figure 2). Use a 100 µl multipipette to mix all the samples 4 x and then transfer 20 µl to row B to perform the -1 dilution. Use a 100 µl multipipette to mix all the samples 4 x and then transfer 20 µl to the next row for the -2 dilution and so on. It is very important to change pipette tips between every dilution performed. Repeat until -5 dilution has been performed (Figure 2).
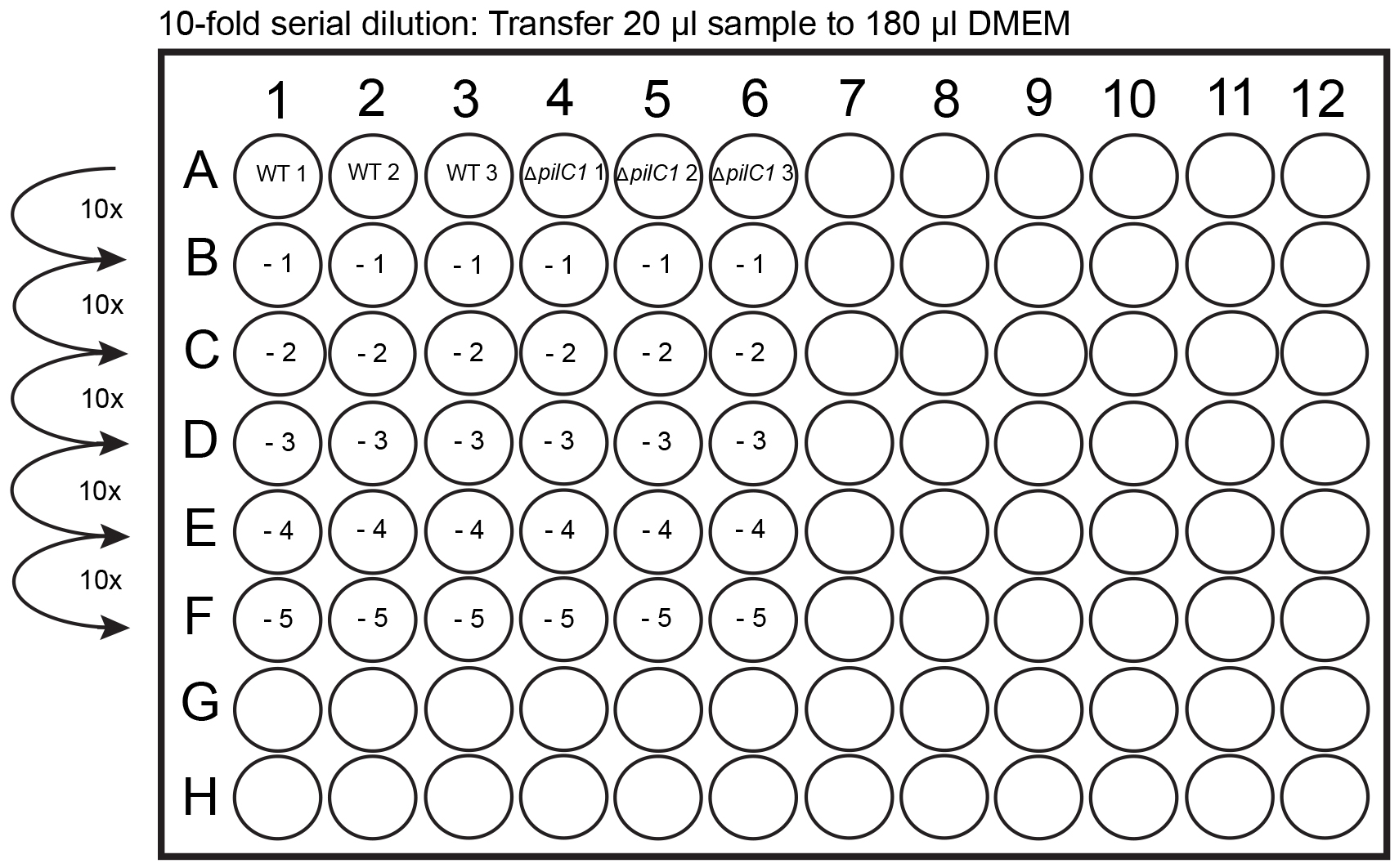
Figure 2. Dilution series performed in a 96-well plate. Add 180 µl of DMEM to -1 to -5 wells. Transfer 20 µl of cell lysates (from row A) to row B containing 180 µl of DMEM. Mix the solution and continue with serial dilutions until the -5 dilution has been completed.
- Spot 50 µl of dilutions -2 (10-2), -3 (10-3), -4 (10-4) and -5 (10-5) for each sample to each square marked on the GC agar plates to acquire countable colonies (Figure 1B). Use the pipette tip or a 1 µl loop to spread the drop, if it does not occur naturally, on the GC agar plates. Allow the drops to dry in the bacterial hood before shifting to an incubator.
- Incubate the GC agar plates in a 37 °C incubator with 5% CO2 overnight.
- Prewarm DMEM. Before the experiment wash the cells gently once and add 800 µl of fresh DMEM. While preparing bacterial cultures, keep the cell culture plate in an incubator at 37 °C with a 5% CO2 environment.
Day 3
- After overnight incubation, count the number of colonies from the dilutions that have between 20-100 colonies.
Data analysis
- To calculate the number of adhered bacteria, multiply the number of colonies with 20 to get the number of bacteria in 1 ml per dilution, since only 50 µl was plated. In order to calculate cfu/ml multiply with 10 for every dilution performed. For example, if 50 colonies were counted from the 5th dilution. 50 x 20 x 105 = 1 x 107. The data can be either presented as adhered bacteria (cfu/ml) or adhered bacteria/cell. Calculate the average of the duplicates/triplicates. Results can be plotted graphically in Microsoft Excel as a column graph.
- Standard deviation can be calculated as well as two-tailed, unpaired Student’s t-test for analysis of statistical significance from three independent adhesion assays.
- Example of data presentation can be observed in Sigurlásdóttir et al. (2017).
Recipes
- GC agar plates (1 L)
To a 1 L flask, add 36 g medium base and fill up to 600 ml of distilled H2O
Autoclave
Add up to 1 L with sterile cold distilled H2O
Add 10 ml of Kellogg’s supplement (Recipe 2) when the temperature is about 60 °C
Pour the agar into Petri dishes and allow the plates to cool down
Store the plates at 4 °C
- Kellogg’s supplement (200 ml)
Dissolve 80 g of D-glucose in 100 ml of distilled H2O
Add 1 g of L-glutamine, 0.1 g of ferric nitrate and 2 ml of 0.2% Cocarboxylase solution (Recipe 3)
Add up to 200 ml with distilled H2O when everything is dissolved
Sterile filter with 0.22 µm vacuum filtration unit and store at 4 °C
- Cocarboxylase solution
0.02 g in 100 ml of distilled H2O, sterile filter with a 0.22 µm filter, store at 4 °C
- 2x trypsin
Dilute to 2x trypsin by adding 5 ml to 20 ml of 1x PBS
- 1% saponin
0.1 g saponin, dissolved in DMEM and filled up to 10 ml
6.Phosphate-based GC liquid medium (500 ml)
Dissolve 7.5 g of protease peptone, 0.5 g of soluble starch, 2 g of potassium phosphate dibasic, 0.5 g of potassium phosphate monobasic and 2.5 g of sodium chloride in 450 ml of distilled H2O. When dissolved, fill up to 500 ml with distilled H2O and autoclave
Acknowledgments
This work was adapted from a protocol we used for studies previously published (Sigurlásdóttir et al., 2017). This work was funded by the Swedish Research Council (Dnr 2006-4112, 2012-2415, 2013-2434), The Swedish Cancer Society and Torsten Söderbergs Stiftelse. The authors declare no conflicts on interest.
References
- de Klerk, N., Saroj, S. D., Wassing, G. M., Maudsdotter, L. and Jonsson, A. B. (2017). The host cell transcription factor EGR1 is induced by bacteria through the EGFR-ERK1/2 pathway. Front Cell Infect Microbiol 7: 16.
- Engman, J., Negrea, A., Sigurlasdottir, S., Georg, M., Eriksson, J., Eriksson, O. S., Kuwae, A., Sjolinder, H. and Jonsson, A. B. (2016). Neisseria meningitidis polynucleotide phosphorylase affects aggregation, adhesion, and virulence. Infect Immun 84(5): 1501-1513.
- Eriksson, J., Eriksson, O. S. and Jonsson, A. B. (2012). Loss of meningococcal PilU delays microcolony formation and attenuates virulence in vivo. Infect Immun 80(7): 2538-2547.
- Helaine, S., Carbonnelle, E., Prouvensier, L., Beretti, J. L., Nassif, X. and Pelicic, V. (2005). PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol 55(1): 65-77.
- Hill, D. J., Griffiths, N. J., Borodina, E. and Virji, M. (2010). Cellular and molecular biology of Neisseria meningitidis colonization and invasive disease. Clin Sci (Lond) 118(9): 547-564.
- Marceau, M., Beretti, J. L. and Nassif, X. (1995). High adhesiveness of encapsulated Neisseria meningitidis to epithelial cells is associated with the formation of bundles of pili. Mol Microbiol 17(5): 855-863.
- Merz, A. J. and So, M. (2000). Interactions of pathogenic Neisseriae with epithelial cell membranes. Annu Rev Cell Dev Biol 16: 423-457.
- Mikaty, G., Soyer, M., Mairey, E., Henry, N., Dyer, D., Forest, K. T., Morand, P., Guadagnini, S., Prevost, M. C., Nassif, X. and Dumenil, G. (2009). Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog 5(2): e1000314.
- Rahman, M., Kallstrom, H., Normark, S. and Jonsson, A. B. (1997). PilC of pathogenic Neisseria is associated with the bacterial cell surface. Mol Microbiol 25(1): 11-25.
- Rudel, T., Scheurerpflug, I. and Meyer, T. F. (1995). Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature 373(6512): 357-359.
- Sigurlásdóttir, S., Engman, J., Eriksson, O. S., Saroj, S. D., Zguna, N., Lloris-Garcera, P., Ilag, L. L. and Jonsson, A. B. (2017). Host cell-derived lactate functions as an effector molecule in Neisseria meningitidis microcolony dispersal. PLoS Pathog 13(4): e1006251.
- Soto, G. E. and Hultgren, S. J. (1999). Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol 181(4): 1059-1071.
- Stephens, D. S. (2009). Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine 27 Suppl 2: B71-77.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sigurlásdóttir, S., Saroj, S. D., Eriksson, O. S., Eriksson, J. and Jonsson, A. (2018). Quantification of Neisseria meningitidis Adherence to Human Epithelial Cells by Colony Counting. Bio-protocol 8(3): e2709. DOI: 10.21769/BioProtoc.2709.
Category
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell-based analysis > Cell adhesion
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









