- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Exosome Transfer in Mammalian Cells by Fluorescence Recovery after Photobleaching
Published: Vol 8, Iss 2, Jan 20, 2018 DOI: 10.21769/BioProtoc.2692 Views: 8746
Reviewed by: Giusy TornilloShuhei OtaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1600 Views

Monitoring of Sperm-Independent Calcium Oscillations in Immature Oocytes of Mice
Sae Horiike [...] Hidehiko Ogawa
Feb 5, 2026 63 Views

High Content In Vitro Survival Assay of Cortical Neurons
Paolo V. Fioretti [...] Manuela Basso
Feb 5, 2026 64 Views
Abstract
During the course of evolution, prokaryote and eukaryote cells have developed elegant and to some extent analogous strategies to communicate with each other and to adapt to their surrounding environment. Eukaryotic cells communicate with each other through direct interaction via juxtracrine signaling and/or by secreting soluble factors. These secreted factors can subsequently act on the cell itself (autocrine signaling) or interact with neighboring (paracrine signaling) and distant (endocrine signaling) cells. The transmission of signals between cells and tissues has been traditionally thought to be regulated by a protein-based signaling system. Typically, proteins destined for secretion into the extracellular milieu by exocytosis contain a canonical secretion-targeting sequence (Théry et al., 2002). However, proteins with a non-continuous and stimulus-dependent secretion, proteins that do not contain a canonical secretion-targeting sequence, and species that might be too labile within the extracellular environment (DNA, mRNA, peptides, metabolites, miRNA and other RNA species), can be secreted in small membranous extracellular vesicles (EVs) in a specific manner (Hagiwara et al., 2014). Exosomes represent one broad class of these secreted membrane vesicles with a diameter of 30-130 nm (Cocucci et al., 2009; Théry et al., 2009; Kowal et al., 2014), which are formed inside the secreting cells in endosomal compartments called multivesicular bodies. Molecules loaded into exosomes as well as the intensity of the exosome transfer between cells are important parameters for the subsequent conditioning of recipient cells. Current knowledge on secretion of exosomes and their internalization in recipient cells remains incomplete. It is known that secretion intensity of exosomes varies according to the cellular type and its physiological state (Garcia et al., 2016). Moreover, the different combination of transmembrane proteins on the surface of exosomes that facilitate the adhesion to the cell-extracellular matrix vary the avidity with which a recipient cell captures exosomes (Hoshino et al., 2015). Here, we have developed an in vitro system by which the transfer of exosomes between cells in co-culture can be quantified using FRAP (‘Fluorescence Recovery After Photobleaching’) technology. This protocol has been used to analyze the effects of exosome transfer of hypoxia inducible factor 1-α (HIF-1α) in Mesenchymal Stem Cells (MSC; HIF-MSC) to Human Umbilical Cord Vein Endothelial Cells (HUVEC) (Gonzalez-King et al., 2017).
Keywords: ExosomesBackground
Exosomes are small lipid bilayer vesicles that function as intercellular messengers. Here we created transgenic mesenchymal stem cell (MSC) lines that express the tetraspanin CD63 exosomal marker fused with Red Fluorescent Protein (RFP) to trace and quantify exosome transfer to Human Umbilical Cord Vein Endothelial Cells (HUVECs) using Fluorescence Recovery After Photobleaching (FRAP) technology. This simple method requires an optical microscope, a light source, and a fluorescent probe. As a first step, a background image of the HUVEC is saved before photobleaching. Next, the light source is focused on the HUVEC, and the RFP fluorophores in this region receive high intensity illumination, which causes their fluorescence lifetime to decrease quickly. Using this approach, red fluorescence recovery would be due to exosome uptake by the photobleached HUVEC. We used this system to examine the exosomal transfer of HIF-MSC to HUVEC as a potential therapeutic approach for angiogenesis.
Materials and Reagents
- Pipette tips (SARSTEDT)
- 0.45 μm sterilize filter filtropur (SARSTEDT, catalog number: 83.1823.100 )
- 25-mm glass coverslips (Thermo Scientific, Menzel-Gläser, Braunscheig, Germany)
- 6-well plates (SARSTEDT, catalog number: 83.3920 )
- Culture-Insert 2-well in μ-dish 35 mm, high ibiTreat: ready to use (IBIDI, catalog number: 81176 )
- Sterile individually packaged 5 ml pipettes (SARSTEDT, catalog number: 86.1253.001 )
- 0.22 μm sterilize filter filtropur (SARSTEDT, catalog number: 83.1826.001 )
- 20 ml syringe (B. Braun Medical, catalog number: 4617207V-02 )
- 50 ml corning tube (SARSTEDT, catalog number: 62.547.254 )
- 15 ml conical tubes (SARSTEDT, catalog number: 62.554.502 )
- Human mesenchymal stem cells from dental pulp
- pCT-CD63-RFP lentiviral vector (System Biosciences, catalog number: CYTO120R-VA-1 )
- HEK293T cells
- Absolute ethanol for analysis (Merck, catalog number: 1009832500 )
- Trypan blue (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061 )
- Trypsin/EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 15400054 )
- DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31885023 )
- Heat inactivated FBS (fetal bovine serum) (Thermo Fisher Scientific, GibcoTM, catalog number: 12484028 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- EGM-2 Bullet Kit (Lonza, catalog number: CC-3162 )
- Puromycin (Sigma-Aldrich, catalog number: P9620 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014 )
- Potassium chloride (KCl) (Merck, catalog number: PX1405 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655 )
- MSC, HIF-MSC and HEK293T culture medium (see Recipes)
- MSC-CD63-RFP and HIF-MSC-CD63-RFP selection medium (see Recipes)
Equipment
- Pipettes (Eppendorf)
- Cell culture hood (any brand)
- Class II Type A2 biological safety hood (any brand)
- Centrifuge (Centrifuge 5804R) (Eppendorf, model: 5804 R )
- Leica TCS SP2 AOBS inverted laser scanning confocal microscope (Leica Microsystems, Heidelberg GmbH, Mnnheim, Germany, http://www.leica-microsystems.com)
- Heating apparatus
Software
- LAS X with FRAP wizard
- ImageJ (https://imagej.softonic.com/descargar?ex=DSK-309.1)
Procedure
In the protocol described below, we analyze the transfer of exosomes from MSC with or without stabilized expression of HIF-1α to HUVECs.
- Production of MSC and HIF-MSC
Cells were transduced with a lentiviral vector coding for HIF-1α and the green fluorescent protein (GFP), named pWPI-HIF-1α-GFP, or a control vector pWPI-GFP (https://www.addgene.org/) as previously described (Ciria et al., 2017).
- Production of MSC-CD63-RFP and HIF-MSC-CD63-RFP
- Retroviral particles were obtained from supernatants of HEK293T packaging cell lines transduced with a vector expressing Red Fluorescent Protein (RFP) fused to the tetraspanin CD63: pCT-CD63-RFP (CYTO120R-PA-1).
- Filter the supernatants with retroviral particles through a 0.45 μm filter.
- Titration of lentiviral supernatants is performed in fresh HEK293T cells with our local Viral Vector production Unit (http://inbiomed.org).
- MSC and HIF-MSC freshly seeded at 5 cells/cm2 are transduced for 8 h at a multiplicity of infection of 10 in MSC or HIF-MSC culture medium.
- After this time, wash off the medium containing lentiviral particles and replace with fresh medium
- Repeat this procedure (Step A5) daily for 3 days.
- MSCs and HIF-MSCs containing the pCT-CD63-RFP vector are selected with puromycin. Culture cells in MSC culture medium with 4 μg/ml puromycin for 24 h. After this time, wash out puromycin and replace with fresh culture medium.
- Evaluate the transduction efficiency in MSC and HIF-MSC by using flow cytometry (cells should be GFP and RFP positive). After selection, purity of transduced populations is routinely higher than 90%.
- Retroviral particles were obtained from supernatants of HEK293T packaging cell lines transduced with a vector expressing Red Fluorescent Protein (RFP) fused to the tetraspanin CD63: pCT-CD63-RFP (CYTO120R-PA-1).
- 24 h co-culture of MSC or HIF-MSC with HUVEC (Figure 1A)
- Sterilize 25-mm glass coverslips by soaking in 70% ethanol for 5 min.
- Air-dry coverslips in the cell culture hood for 10 min and then place into 9 cm2 wells.
- Seed a total of 15 x 103 MSC-CD63-RFP or HIF-MSC-CD63-RFP cells and 105 HUVEC in co-culture or separately onto the 25-mm sterilized glass coverslips. Co-culture medium is a 1:1 mixture of HUVEC and MSC growth medium.
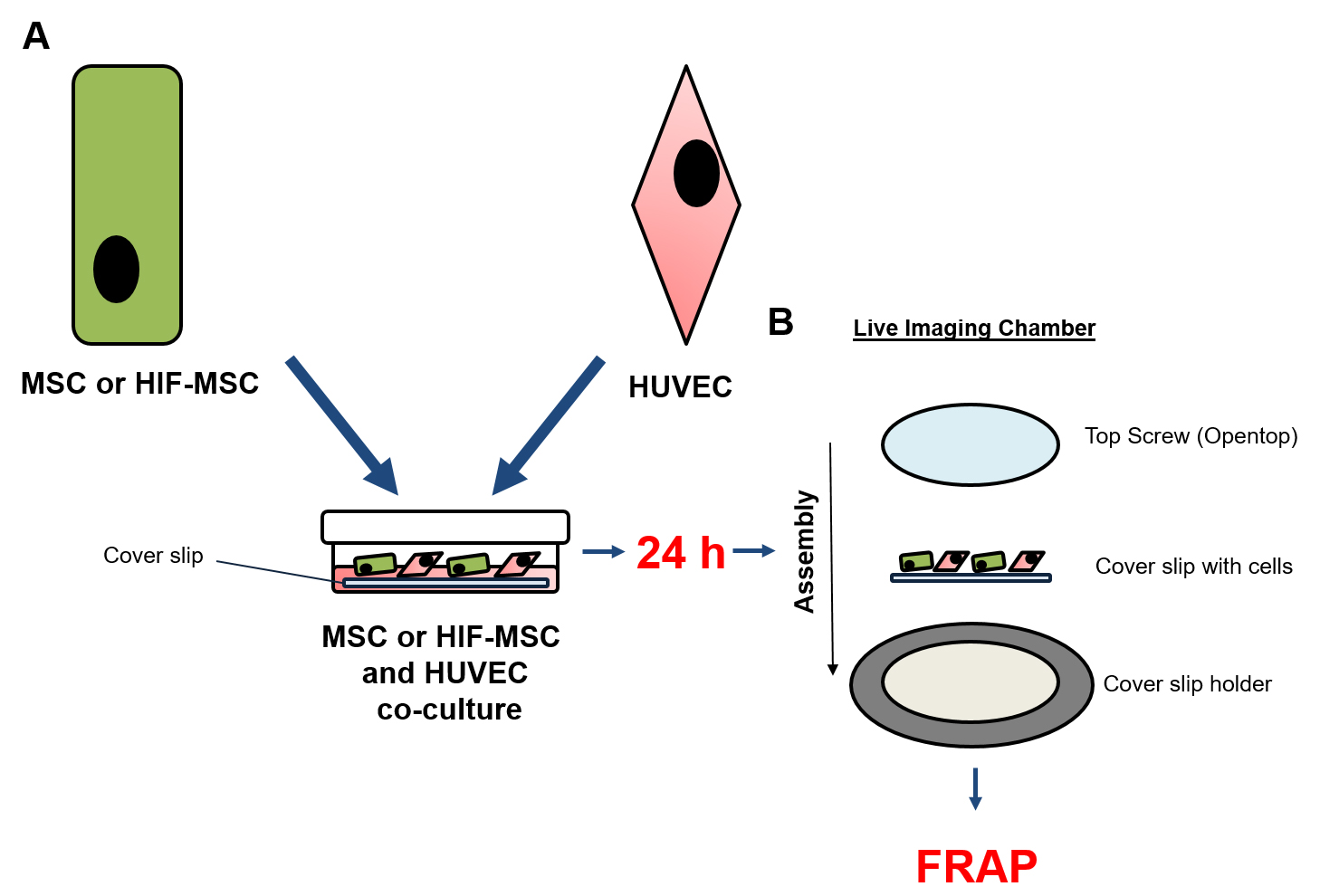
Figure 1. Co-culture scheme. A. Co-culture of HUVEC and MSC or HIF-MSC onto cover slip; B. Montage of co-culture cover slip into Live Imaging Chamber.
- Sterilize 25-mm glass coverslips by soaking in 70% ethanol for 5 min.
- FRAP
- After 24 h of co-culture, filter conditioned medium through a 0.22 μm filter to remove cell debris and large EVs, and then add the filtered medium back into the same well. Most of tetraspanin CD63 is concentrated in the exosomes; however, there are EVs of various sizes that also contain CD63. Thus, to reduce the presence of vesicles other than exosomes, a selection is made by size filtering the culture medium after 24 h of culture.
- Mount co-cultures in live imaging chambers and observe by confocal microscopy (Figure 1B). As HUVEC are not fluorescently labeled, detection of red fluorescence in these cells could only result from incorporation of red nanovesicles secreted from MSC-CD63-RFP and HIF-MSC-CD63-RFP cells that emit both green and red fluorescence.
- A single HUVEC is photo-bleached with a 460-480 nm laser as described in Gonzalez-King et al., 2017. Of note, photobleaching conditions have to be established for each cell type and for the fluorescent molecules used.
- Always take images in the same ‘z’ plane of the cell. Thus, it is recommended that before starting image, ensure that red fluorescence (due to red fluorescent exosomes uptake during the 24 h of co-culture) is no longer present. To do this, examine the photobleached cells in different ‘z’ planes before imaging.
- After photo-bleaching of a single HUVEC, two-dimensional pseudo-color images (255 color levels) are acquired using a 40x Plan-Apochromat-Lambda Blue 1.4 NA oil objective with a size of 1,024 x 1,024 pixels and Airy 1 pinhole diameter, at 10 min intervals during 3 h. The excitation wavelengths for fluorochromes are 488 nm (argon laser) for the detection of GFP fluorescence and 561 nm (DPSS laser) for detection of DsRed. To avoid variability, images must be taken under the same conditions established at the beginning of the experiment. During the analysis, maintain slides at 37 °C with a heating apparatus supplied with a 95% air/5% CO2 humidified gas mixture.
- After 24 h of co-culture, filter conditioned medium through a 0.22 μm filter to remove cell debris and large EVs, and then add the filtered medium back into the same well. Most of tetraspanin CD63 is concentrated in the exosomes; however, there are EVs of various sizes that also contain CD63. Thus, to reduce the presence of vesicles other than exosomes, a selection is made by size filtering the culture medium after 24 h of culture.
Data analysis
- Fluorescence recovery analysis was done using ImageJ. To do this, only red/green composite fluorescence images (without the transmitted light picture) of the Regions Of Interest (ROI) from inside the photobleached HUVEC were measured from the images before and after the photobleaching, and from each image after the photobleaching for 3 h as described in Figure 2.
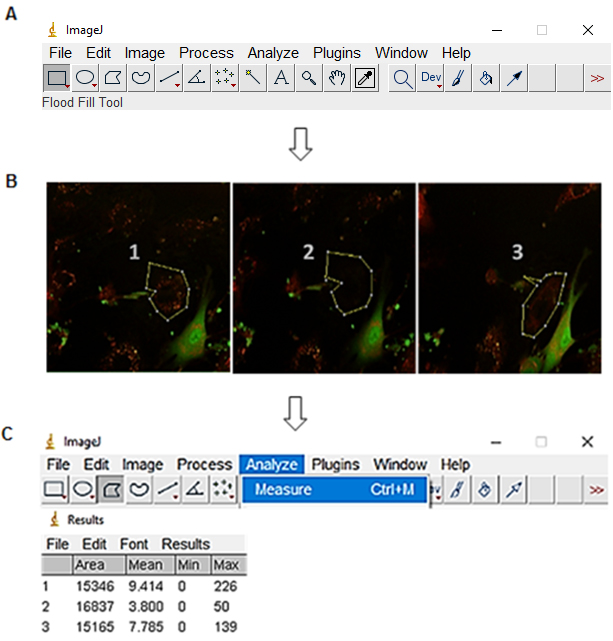
Figure 2. Fluorescence intensity measurement of the ROI with ImageJ. A. Open ImageJ and select Polygons selections option. B. Select the region with red fluorescence of the cell that has been photobleached. C. Select analyze and measure to give a numerical value to the fluorescence of the ROI.
To measure the fluorescence recovery, 100% of fluorescence recovery was considered the fluorescence intensity of the imaged HUVEC before photobleaching. The fluorescence intensity of the first image of HUVEC after photobleaching was considered as the percentage of residual fluorescence. Subsequently, the fluorescence intensity of images captured up to 3 h after photobleaching was measured and represented as a % of recovery regarding the image before photobleaching the HUVEC (Figure 3).
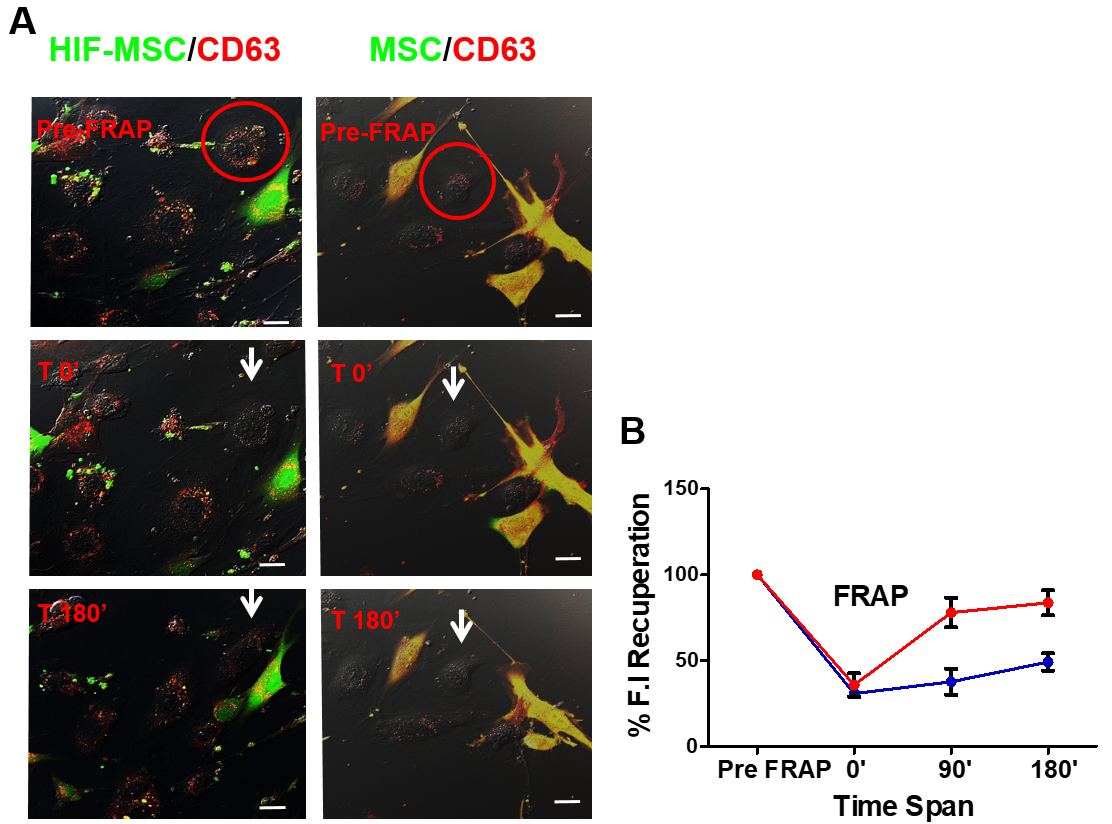
Figure 3. Recovery of fluorescence after photobleaching in Human Umbilical Vein Endothelial Cells (HUVECs) co-cultured with MSC-CD63-red fluorescence protein (RFP) or HIF-MSC-CD63-RFP cells. A. White circle, photobleached cell; arrow, photobleached cell after 0 and 180 min of bleaching; pre-fluorescence recovery after photobleaching (FRAP), confocal image of cells before photobleaching; T0’, confocal image of the cells after photobleaching at time 0’; T180’, confocal image of the cells 180 min after photobleaching. Images were taken at 40x magnification. B. Percentage of the fluorescence recovery of HUVEC bleached at timepoints 0, 90, and 180 after photobleaching, cultured with MSC-CD63-RFP cells (black plotted line) or HIF-MSC-CD63-RFP cells (grey plotted line). Results are expressed relative to T0’. **, P < .01; ***, P < .001. n = 3 in every experiment (Abbreviations: FRAP, fluorescence recovery after photobleaching; HIF, hypoxia-inducible factor; HIF-MSC, mesenchymal stem cells overexpressing HIF-1α; MSC, mesenchymal stem cell).
- Results in the example shown in Figure 1 show a higher percentage of red fluorescence recuperation after 3 h of HUVEC co-cultured with HIF-MSC than with MSC. This finding suggests that the transference of exosomes was higher from HIF-MSC to HUVEC than in co-cultures where MSC was used.
Notes
In this work, we analyzed exosome transfer from one cell type, subjected or not to overexpression of HIF-1α, to another cell type in co-culture. This protocol should be applicable for analysis of exosome transfer in other cell types; however, the correct conditions of the experiment should be established in each case.
Recipes
- MSC, HIF-MSC and HEK293T culture medium
DMEM with 10% of heat-inactivated FBS and 5% of penicillin/streptomycin
HUVEC culture medium was EGM-2 Bullet Kit
- MSC-CD63-RFP and HIF-MSC-CD63-RFP selection medium
DMEM with 10% of heat-inactivated FBS, 5% penicillin/streptomycin and 4 μg/ml puromycin
Acknowledgments
This work was supported in part by grants from the Instituto de Salud Carlos III (PI16/00107, DTS15/00079, DTS17/0067), RETICS RD16/0011/0004 to PS co-funded by FEDER ‘una manera de hacer Europa’. It was also supported by the Regenerative Medicine Program of Instituto de Salud Carlos III and the Valencian Community to Centro de Investigación Principe Felipe. H. G-K acknowledges a predoctoral fellowship associated with the Hecatos project (FP7-HEALTH-2013-INNOVATION). This work has been published in Stem Cells, where the protocol was used for the first time (Gonzalez-King et al., 2017). Authors declare no conflict of interest.
References
- Ciria, M., Garcia, N. A., Ontoria-Oviedo, I., Gonzalez-King, H., Carrero, R., De La Pompa, J. L., Montero, J. A. and Sepulveda, P. (2017). Mesenchymal stem cell migration and proliferation are mediated by hypoxia-inducible factor-1α upstream of Notch and SUMO pathways. Stem Cells Dev 26(13): 973-985.
- Cocucci, E., Racchetti, G. and Meldolesi, J. (2009). Shedding microvesicles: artefacts no more. Trends Cell Biol 19(2): 43-51.
- Garcia, N. A., Moncayo-Arlandi, J., Sepulveda, P. and Diez-Juan, A. (2016). Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc Res 109(3): 397-408.
- Gonzalez-King, H., García, N. A., Ontoria-Oviedo, I., Ciria, M., Montero, J. A., Sepúlveda, P. (2017). Hypoxia inducible factor-1α potentiates jagged 1-mediated angiogenesis by mesenchymal stem cell-derived exosomes. Stem Cells 35(7):1747-59.
- Hagiwara, K., Ochiya, T. and Kosaka, N. (2014). A paradigm shift for extracellular vesicles as small RNA carriers: from cellular waste elimination to therapeutic applications. Drug Deliv Transl Res 4(1): 31-37.
- 6. Hoshino, A., Costa-Silva, B., Shen, T. L., Rodrigues, G., Hashimoto, A., Tesic Mark, M., Molina, H., Kohsaka, S., Di Giannatale, A., Ceder, S., Singh, S., Williams, C., Soplop, N., Uryu, K., Pharmer, L., King, T., Bojmar, L., Davies, A. E., Ararso, Y., Zhang, T., Zhang, H., Hernandez, J., Weiss, J. M., Dumont-Cole, V. D., Kramer, K., Wexler, L. H., Narendran, A., Schwartz, G. K., Healey, J. H., Sandstrom, P., Labori, K. J., Kure, E. H., Grandgenett, P. M., Hollingsworth, M. A., de Sousa, M., Kaur, S., Jain, M., Mallya, K., Batra, S. K., Jarnagin, W. R., Brady, M. S., Fodstad, O., Muller, V., Pantel, K., Minn, A. J., Bissell, M. J., Garcia, B. A., Kang, Y., Rajasekhar, V. K., Ghajar, C. M., Matei, I., Peinado, H., Bromberg, J. and Lyden, D. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527(7578): 329-335.
- Kowal, J., Tkach, M. and Thery, C. (2014). Biogenesis and secretion of exosomes.CurrOpin Cell Biol 29: 116-125.
- Théry, C., Ostrowski, M. and Segura, E. (2009). Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9(8): 581-593.
- Théry, C., Zitvogel, L. and Amigorena, S. (2002). Exosomes: composition, biogenesis and function. Nat Rev Immunol 2(8): 569-579.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
González-King, H., García, N. A., Ciria, M., Gascón, S. T., Sánchez, R. S., Grueso, H., Gómez, M., Cabezuelo, R. M., Cava, V. L. and Sepúlveda, P. (2018). Analysis of Exosome Transfer in Mammalian Cells by Fluorescence Recovery after Photobleaching. Bio-protocol 8(2): e2692. DOI: 10.21769/BioProtoc.2692.
Category
Cell Biology > Cell imaging > Fluorescence
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











