- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Micro-computed Tomography to Visualize Vascular Networks in Maize Stems
Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2682 Views: 10364
Reviewed by: Annis Elizabeth RichardsonRenate WeizbauerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Imaging of the Shoot Apical Meristem of Intact, Soil-Grown, Flowering Arabidopsis Plants
Gabriele Bradamante
Jun 20, 2024 2272 Views

Closed Systems to Study Plant–Filamentous Fungi Associations: Emphasis on Microscopic Analyses
Vasiliki Skiada and Kalliope K. Papadopoulou
Feb 20, 2025 2854 Views

Micrografting Technique of Hevea brasiliensis In Vitro Plantlets
Florence Dessailly [...] Julie Leclercq
Feb 20, 2025 1479 Views
Abstract
Plant vascular systems in the stem connect roots with aerial organs to move solutes containing minerals, nutrients as well as signaling molecules, and therefore, they play pivotal roles in plant growth and development. However, stem vascular systems, especially in crop species, have been poorly described since they are deeply embedded in the tissue. Here we describe a protocol to utilize micro-computed tomography (micro-CT) scanning to visualize vascular networks in the maize stem. The protocol covers sample fixation and staining with contrasting reagents, data acquisition using micro-CT, reconstructing three-dimensional (3D) models of stem inner structures and extraction of vascular networks from the model. This protocol can be easily applied to various types of species and organs/tissues.
Keywords: Micro-CT scanningBackground
Monocot stems have a characteristic vascular network in which veins remain separate and independent. Despite its importance to supporting growth and development, the pattern of vascular networks in monocot stems has been poorly studied. Visualization of stem vascular networks is quite challenging because veins are deeply embedded in tissues. Conventional tissue sectioning can be applied to observe the networks, however, it is a laborious and time-consuming process which requires observation of hundreds of sections. In addition, the size of crop stems, which is much larger than the field of view under the microscopes, makes it difficult to capture the whole system.
Recently, we reported that maize transcription factor BEL1-like homeobox (BLH) 12 and BLH14 play important roles in the stem development and vein network formation (Tsuda et al., 2017). To obtain a comprehensive view of the vascular systems, we adopted the micro-CT scanning described previously and optimized it for maize stems (Metscher, 2009a and 2009b, Degenhardt et al., 2010, Staedler et al., 2013, Gignac et al., 2016). By combining this method with image analyses, we were able to reconstruct 3D models of inner stem structures in an efficient and reliable manner. This protocol can be used to visualize inner structures such as veins in various species and tissues.
Materials and Reagents
- 50 ml Polypropylene tube (Greiner Bio One International, catalog number: 227261 )
- Kimwipes
- 1.5 ml micro-tube (Eppendorf, catalog number: 0030125150 )
- Filter foams MOLTOFILTER MF-13 thickness 5 mm (INOAC Corp.)
- Maize (B73)
- Formalin (Wako Pure Chemical Industries, catalog number: 061-00416 )
- Acetic acid (Wako Pure Chemical Industries, catalog number: 017-00256 )
- Ethanol (Wako Pure Chemical Industries, catalog number: 057-00451 )
- Lugol stock solution
- Potassium iodide (Wako Pure Chemical Industries, catalog number: 166-03971 )
- Iodine (Wako Pure Chemical Industries, catalog number: 094-05421 )
- Fixative FAA solution (see Recipes)
- Iodine staining (Degenhardt et al., 2010) (see Recipes)
- 25% Lugol working solution (see Recipes)
Equipment
- X-ray micro-CT imaging system (Comscantechno, model: ScanXmate-E090S105 ) (Figure 1)
- X-ray tube: Microfocus X-ray source (Hamamatsu Photonics K.K., model: L9421-02 )
- Detector: Flat panel detector (Varex Imaging, model: PaxScan 1313DX )
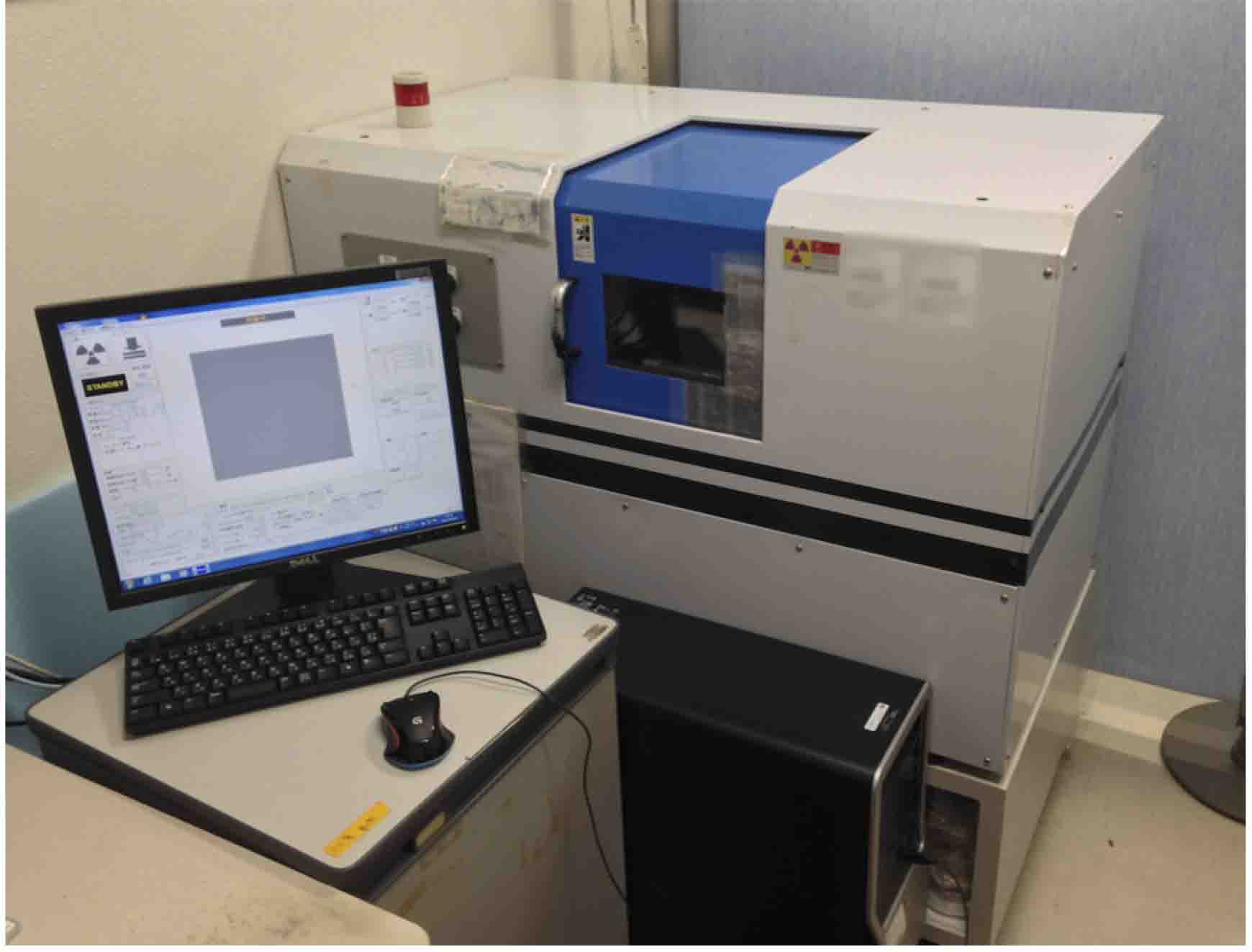
Figure 1. X-ray micro-CT imaging system used in this protocol
- X-ray tube: Microfocus X-ray source (Hamamatsu Photonics K.K., model: L9421-02 )
Software
- coneCTexpress (built in the micro-CT imaging system. Comscantechno Co. Ltd., Kanagawa, Japan)
- OsiriX MD v8.5 (http://www.osirix-viewer.com, Pixmeo SARL, Swiss)
- Imaris 8.2 (http://www.bitplane.com, Bitplane, UK)
- Adobe Premiere Pro CC (http://www.adobe.com, Adobe, US)
Procedure
- Sample fixation and staining with contrasting reagents
- Maize stem dissection and fixation
- Grow maize plants for 6-8 weeks. Our growth condition is at 26 °C under 14-h-light and 10-h-dark condition in the green house. At 6 weeks, the floral transition of the shoot apical meristem has already occurred and the tassel primordium is developing.
- Dissect maize shoot into ~10 cm long sections (Figure 2). Make sure to include the shoot apex in this region. As internodes become shorter toward the apex, we can assume the approximate position of the shoot apex.
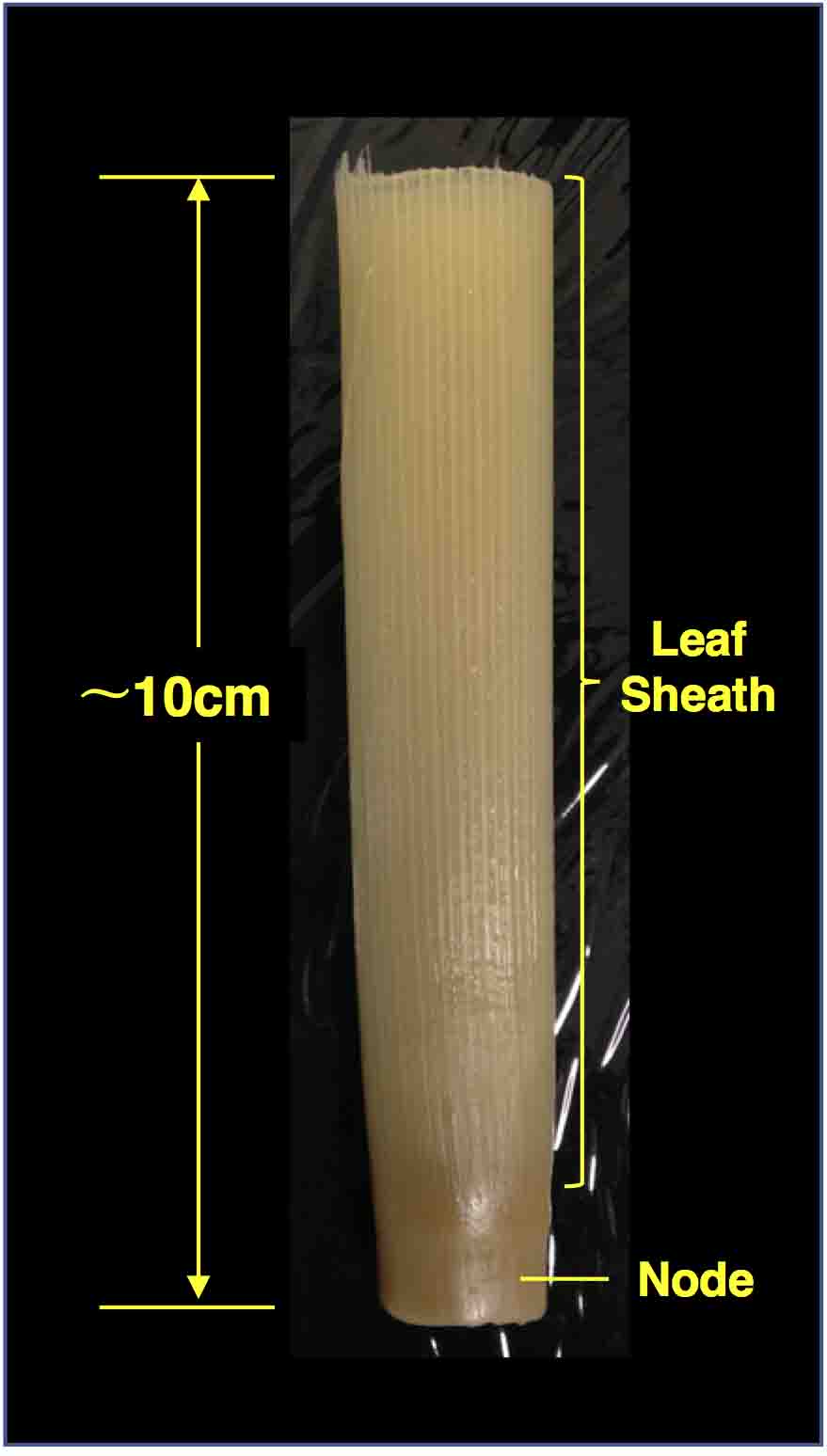
Figure 2. A maize stem sample fixed in FAA - Put the sample into a 50 ml conical tube and fill the tube with FAA solution (see Recipes).
- Slowly vacuum the sample until small air bubbles begin to come out from the samples, then seal the chamber. After 15 min, slowly release the vacuum pressure.
- After releasing the vacuum, replace the solution with new FAA. Vacuum the sample for 15 min again.
- Replace the solution with new FAA and store the sample at 4 °C for at least 1 week. g. Replace FAA with 70% EtOH and rotate the sample tube at 4 °C for 3 h. Replace 70% EtOH with new 70% EtOH. Store the sample in 70% EtOH at 4 °C until observation.
Note: FAA and 70% EtOH containing formaldehyde must be disposed of according to regulations of your institute/university and local governments.
- Grow maize plants for 6-8 weeks. Our growth condition is at 26 °C under 14-h-light and 10-h-dark condition in the green house. At 6 weeks, the floral transition of the shoot apical meristem has already occurred and the tassel primordium is developing.
- Sample staining with contrasting reagents
- Replace 70% EtOH with 35% EtOH and incubate for 2 h at room temperature.
- Discard 35% EtOH and incubate the samples in distilled water at room temperature for 1.5 h. Repeat this step two more times with fresh distilled water.
- Replace the water with 25% Lugol solution (see Recipes) and vacuum for 15 min. Stain the sample for 6 days at room temperature in the dark.
Note: Staining duration depends on the type of tissues and their size. Check the staining before detailed observation (please see Step C2). For maize stems, it takes 6 days (Figure 3).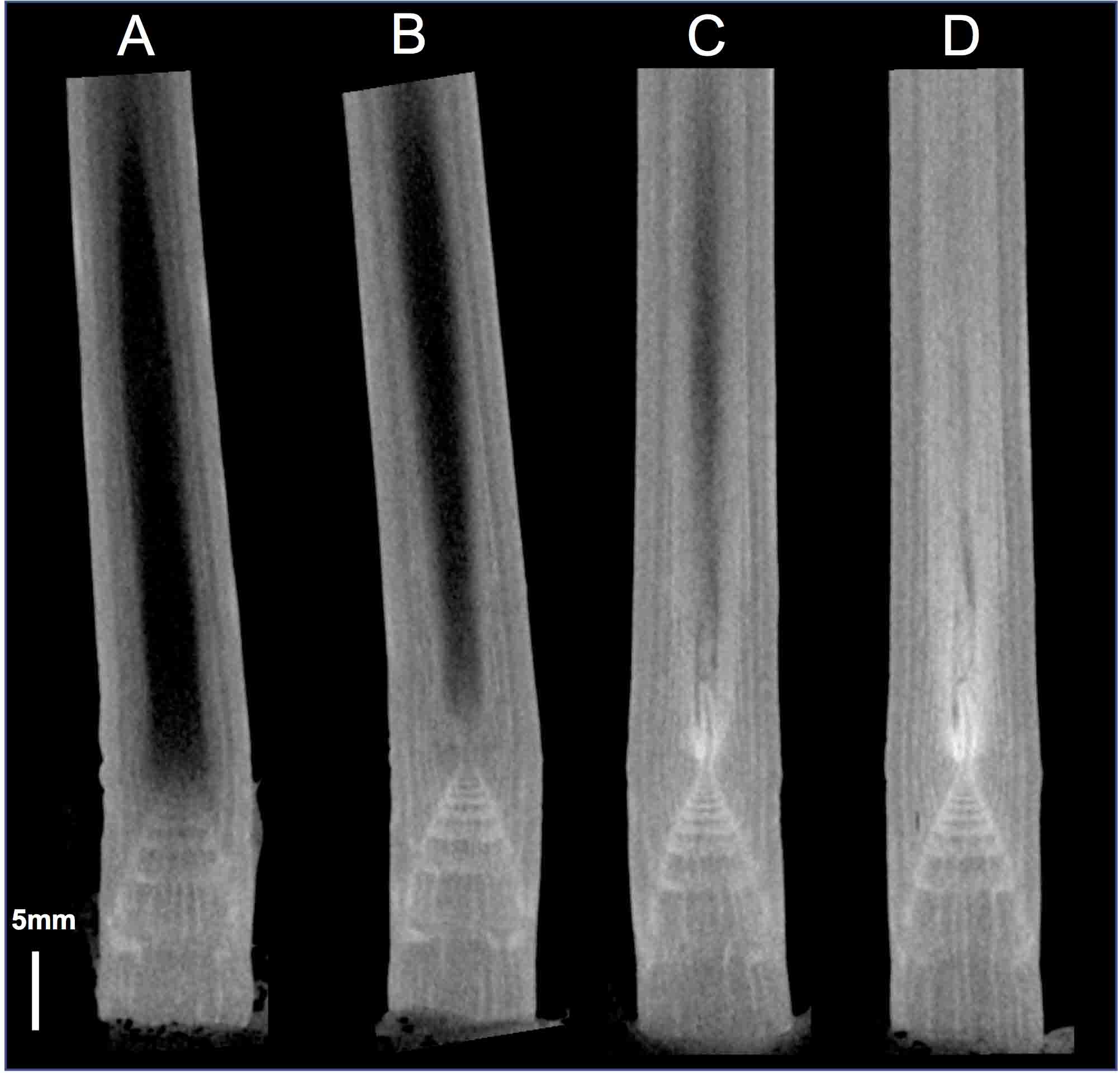
Figure 3. Time-course observation of a maize stem stained with Lugol solution. A maize stem stained in 25% Lugol solution for 1 day (A), 2 days (B), 4 days (C) and 6 days (D) was scanned using micro-CT. These images were processed and displayed in OsiriX. Scale bar = 5 mm.
- Replace 70% EtOH with 35% EtOH and incubate for 2 h at room temperature.
- Maize stem dissection and fixation
- Mounting samples
- Place wet Kimwipes and filter foams at the bottom of 50 ml conical tube.
- Put the stained maize stem in the center of the tube. Make sure to insert the sample so that the lower side of the sample comes to the bottom of the tube (Figure 4A).
Notes:- Softwares used in the downstream data analysis (OsiriX and Imaris) display the imported data upside down. To avoid cumbersome data processing to correct this, we simply mount the samples upside down.
- As Lugol staining is rapidly released from tissue into water, just remove excess solution with paper towels. Do not rinse the sample with water.
- Samples will be rotated on the stage in micro-CT equipment during the data acquisition. It is important to place samples in the center of the stage to obtain high resolution images.
- Softwares used in the downstream data analysis (OsiriX and Imaris) display the imported data upside down. To avoid cumbersome data processing to correct this, we simply mount the samples upside down.
- Place filter foams and wet Kimwipes on the other end of the sample and close the tube.
Note: Maize stems become softer after Lugol staining. Handle the sample gently to avoid deformation. At the same time, make sure to place enough wet Kimwipes and filter foams so that the sample doesn’t move in the tube. The movement of samples during data acquisition will result in a blurred image. Also, moisture in the tube is important to avoid sample drying and deformation.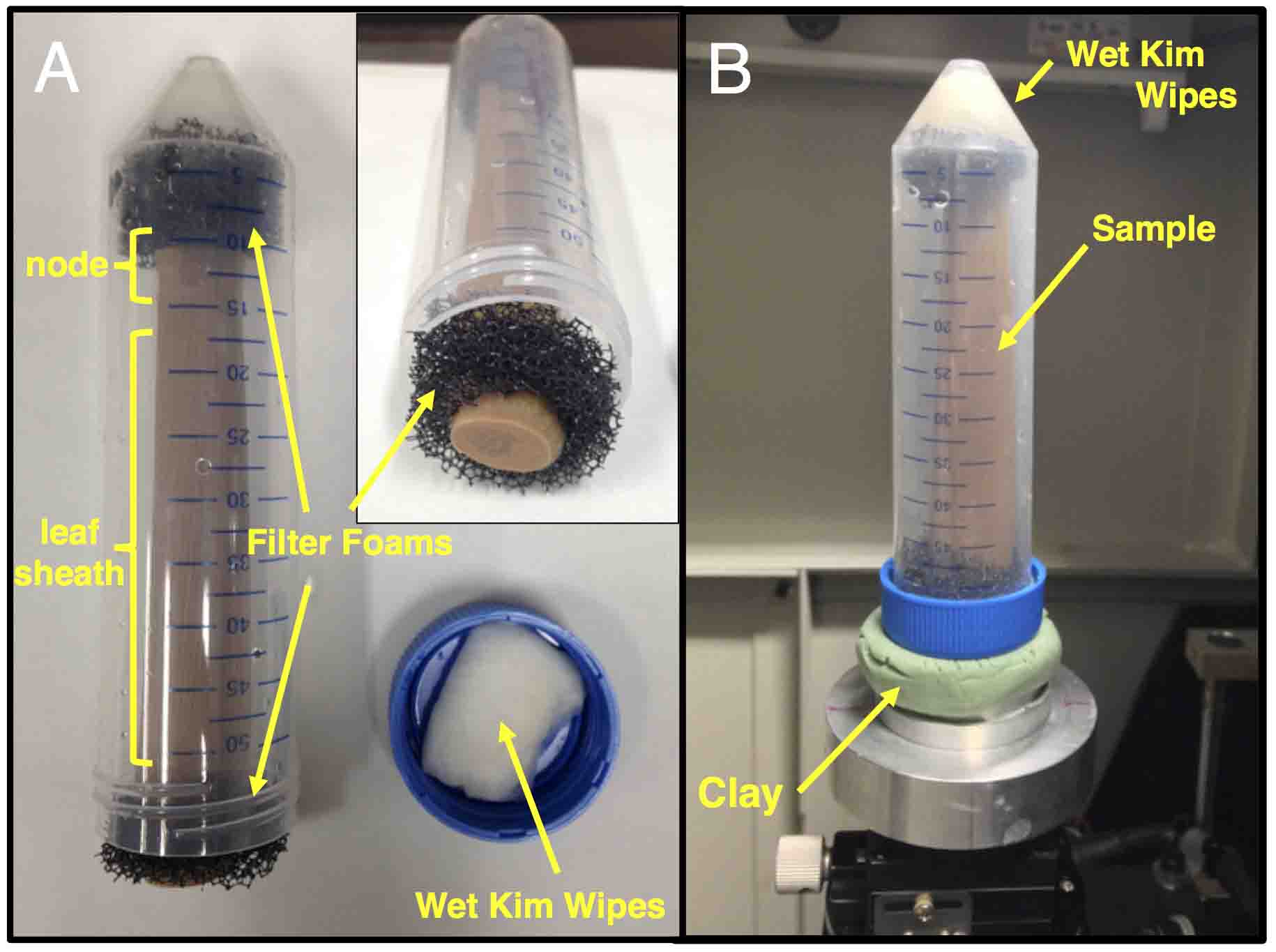
Figure 4. Mounting a maize stem for micro-CT scanning. A. The stained maize stem was mounted in a 50 ml polypropylene tube using wet Kimwipes and filter foams. B. The tube was attached on the stage in micro-CT scanner with a piece of clay.
- Place wet Kimwipes and filter foams at the bottom of 50 ml conical tube.
- Data acquisition
- Put a piece of clay on the sample stage in the micro-CT equipment and fix the sample tube tightly on the clay (Figure 4B).
- Set the voltage at 90 kV and the current at 85 µA. Close the door of the equipment and turn on the X-ray. Now you can see the sample displayed in the fluoroscopic image monitor. To make the stem inner structure visible in the monitor, narrow down the luminance window of the fluoroscopic image. If the sample is stained enough, you can see the conical shape of the shoot apex in the monitor at this point (Figure 5). Determine the position of the shoot apex.
- Check the sample by rotating the stage. Adjust the tube angle so that the sample stands upside down in the center at a right angle to the stage (Figure 5A).
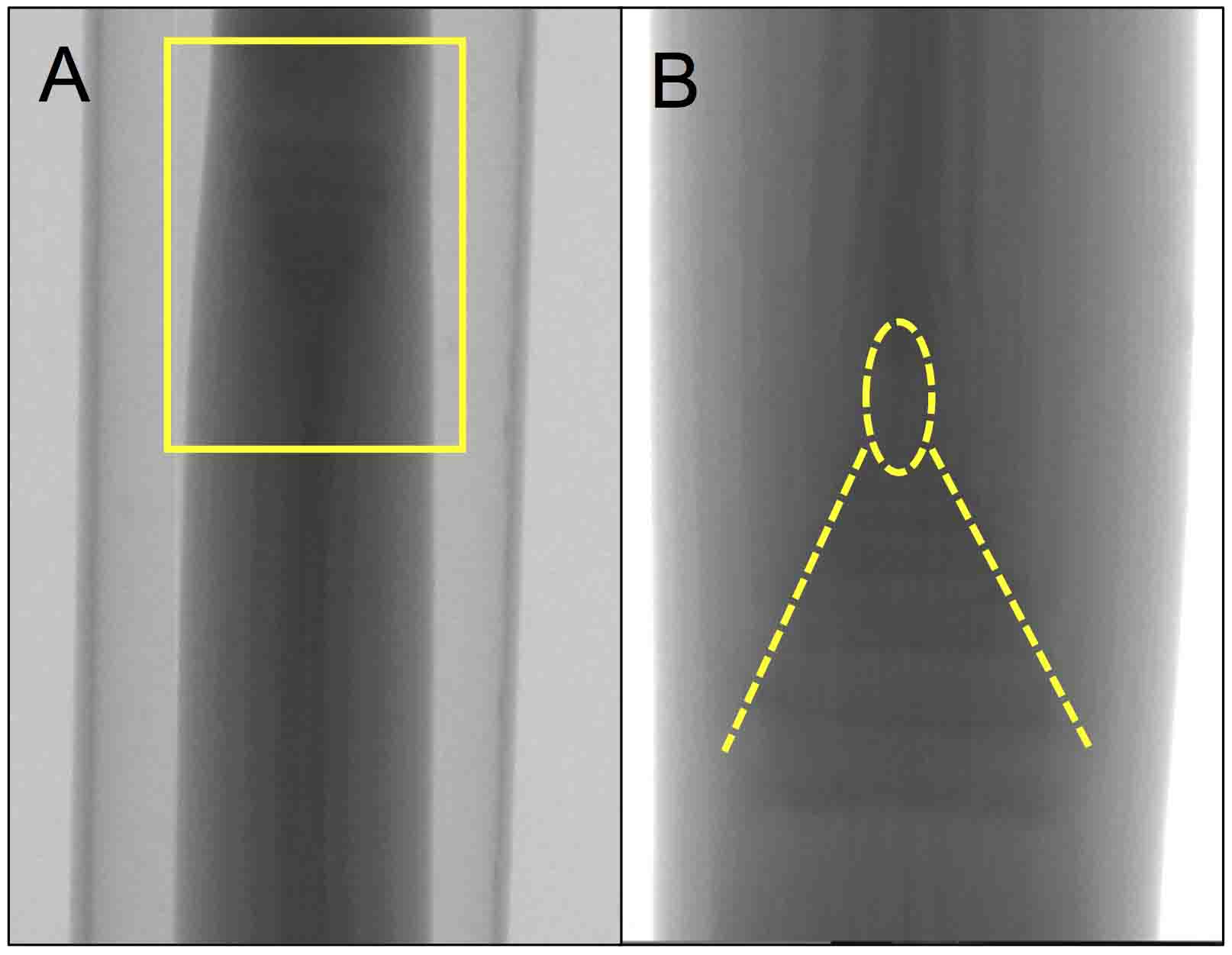
Figure 5. X-ray fluoroscopic imaging before scanning. A. The stem sample mounted on the stage. Note that the stem sample is upside down. A yellow box represents region magnified in (B). B. A magnification of the boxed region in (A). Dashed lines and circle depict a young stem and tassel primordium, respectively. The image was rotated 180° from (A). - Return the luminance window to its full range.
Note: We adjusted the luminance window just to make the sample visible on the monitor. In data acquisition, the luminance window should be the full range. - Determine the scanning area and set scanning parameters (e.g., Table S1) depending on the purpose of your experiment. It is a trade-off between data acquisition area and resolution as shown in (Figure 6).
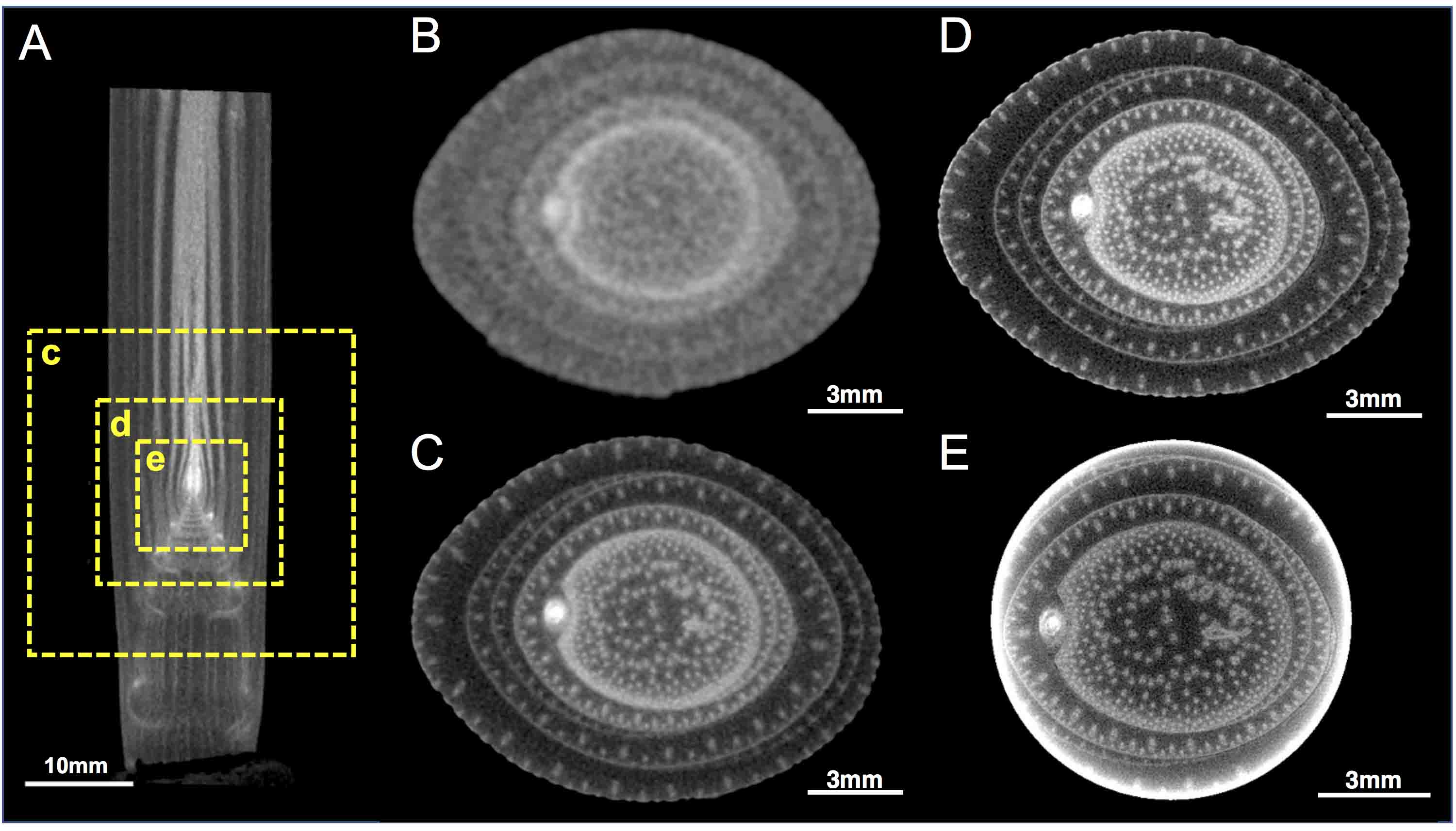
Figure 6. Maize stem images scanned using micro-CT at various resolution. A. A vertical section of the stem. Dashed boxes (c) (d) and (e) represent the magnified regions in (C), (D) and (E), respectively. B-E. Transverse sections of the stem. The resolutions are 67.7, 30.0, 17.0 and 10.0 µm/pixel in (A and B), (C), (D) and (E), respectively. The detailed settings in the scanning are provided in Table S1. - Scan the samples.
Note: When the sample size is larger than the area of data acquisition, the surrounding tissue outside of the field of view causes various noise (Figure 6E). If higher resolution is necessary, trimming the sample to remove surrounding tissue/organs will reduce noise in the data (see below). - If necessary, remove leaves and stems to expose the part of your interest. To mount smaller samples, we use micro-tubes of appropriate size. Put wet Kimwipes and filter foams to fix the sample inside of the tube (Figure 7).
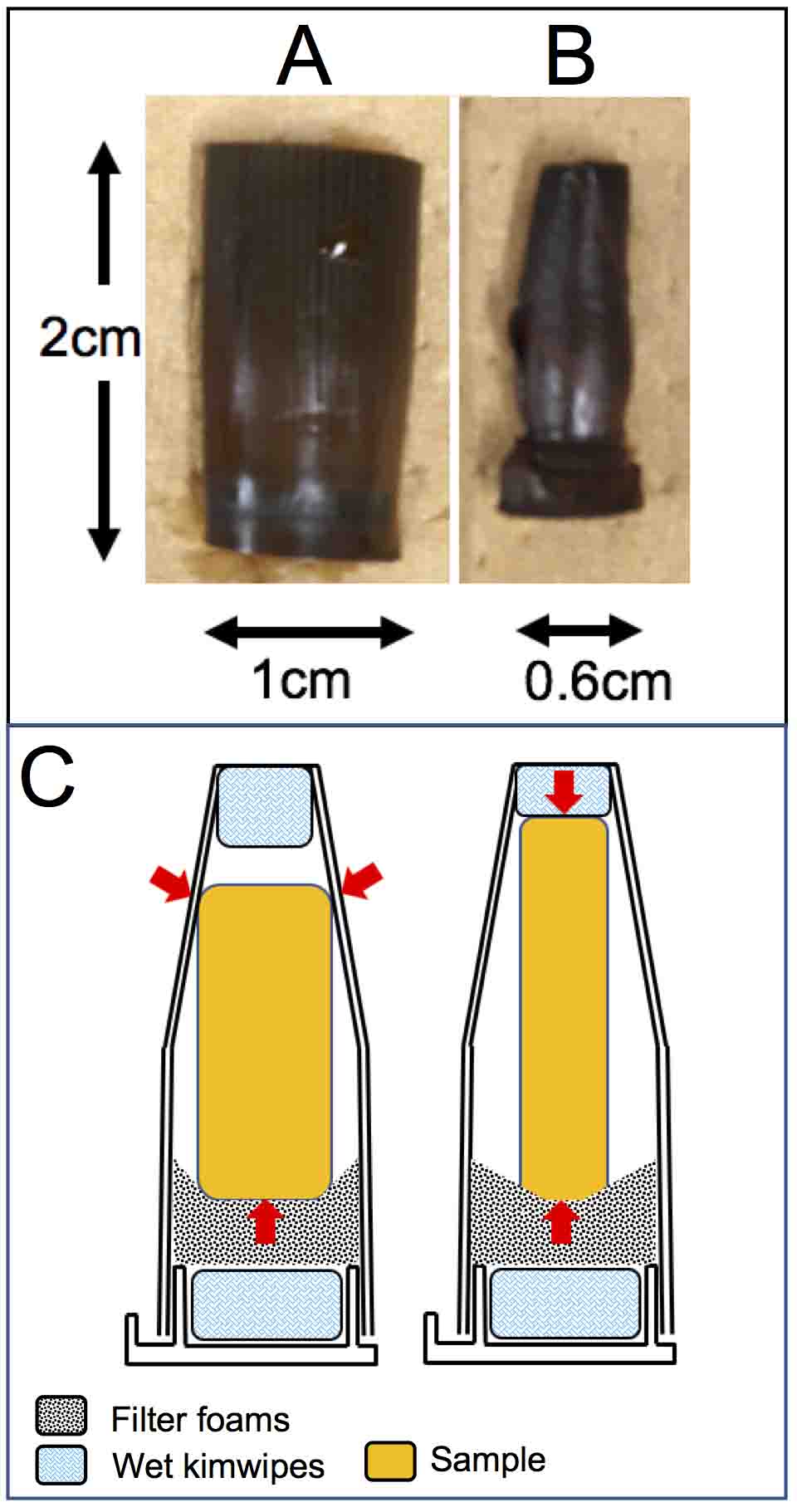
Figure 7. Mounting trimmed maize stems in 1.5 ml tubes. A. Tenth and eleventh leaves from the tassel were removed from the stem sample shown in Figure 2. B. Ninth and eighth leaves were further removed. C. Diagrams showing the trimmed maize stem mounted in 1.5 ml tubes. Red arrows indicate supporting points of samples. - Scan the samples with the setting for higher resolution (Table S2). Figure 8 shows that trimming the samples greatly reduces the noise and enables to increase the resolution.
Note: The resolution of micro-CT images could be further improved by increasing frame average and number of projections (and hence, scanning time as well) (Table S2 and Figure 9).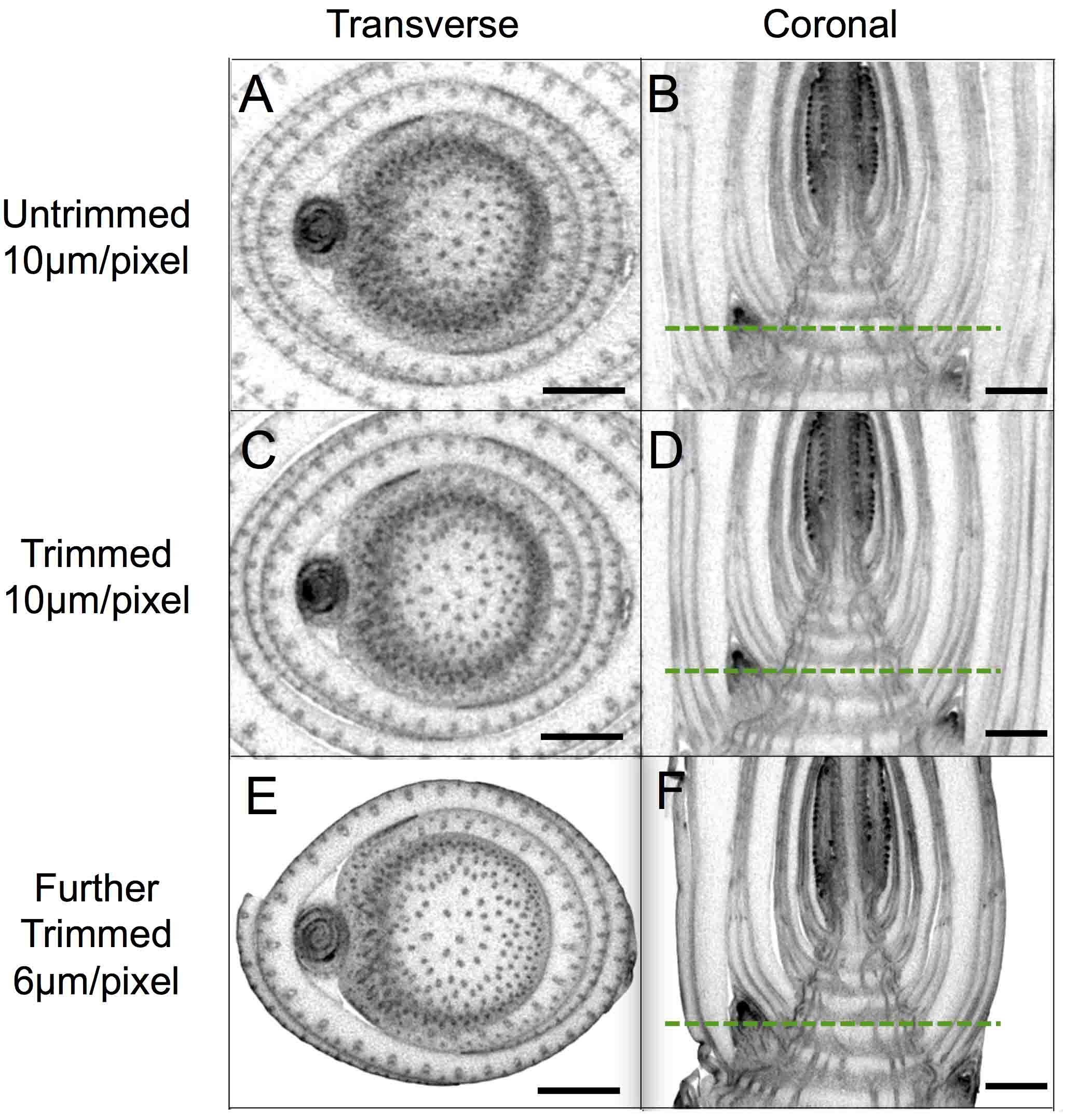
Figure 8. Micro-CT images at improved resolution achieved by trimming samples. A, C and E. Transverse sections of untrimmed (A) and trimmed (C and E) stem. B, D and F. Longitudinal sections of untrimmed (B) and trimmed (D and F) stem. Dashed green lines represent planes shown in transverse sections. Stem samples in Figures 2, 7A and 7B correspond to the micro-CT images in (A and B), (C and D) and (E and F), respectively. Scale bars = 1 mm.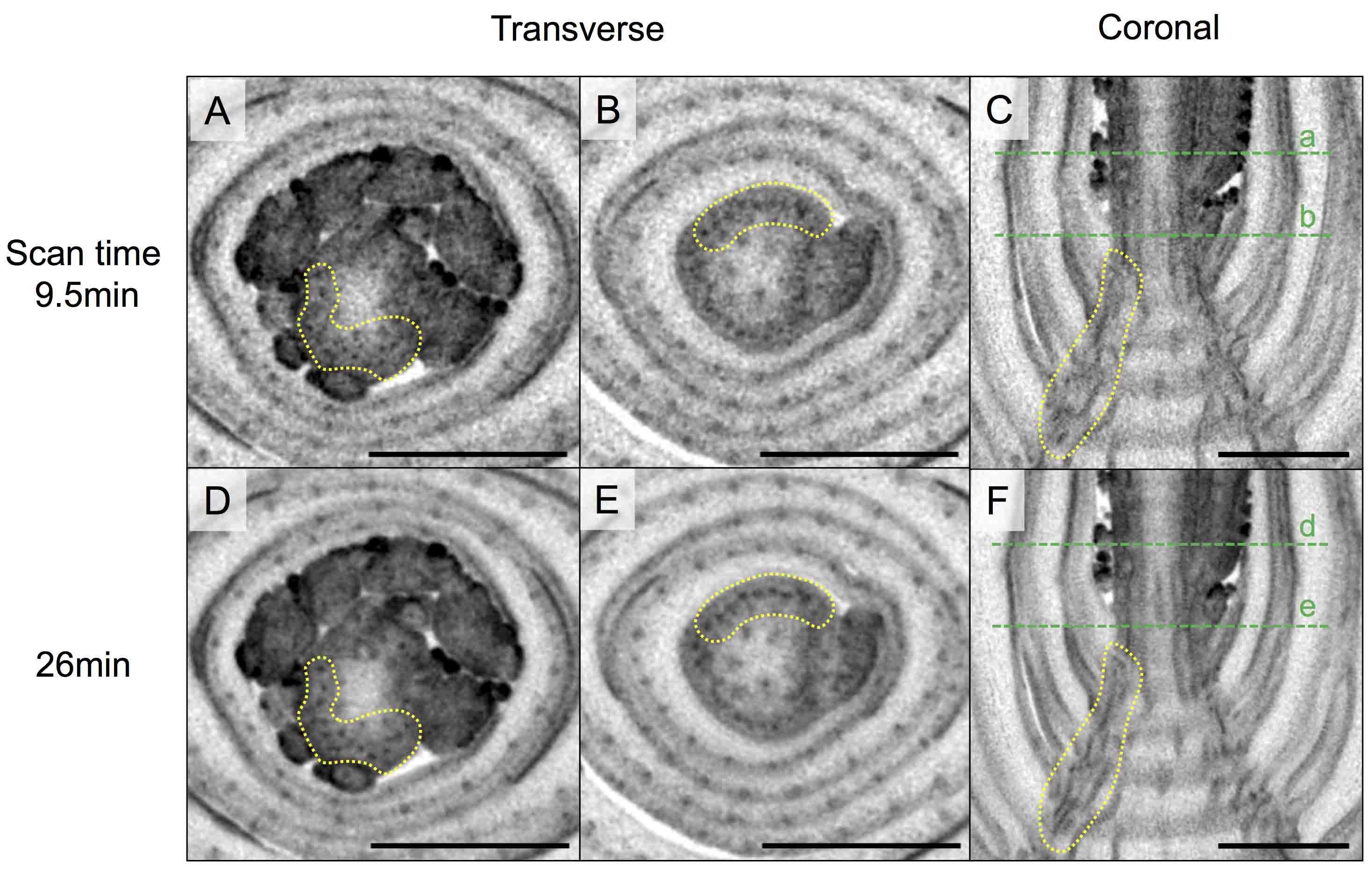
Figure 9. Micro-CT images at further improved resolutions achieved by increasing frame average and number of projections. Micro-CT images of the maize shoot apex scanned for 9.5 min (A-C) and 26 min (D-E). A, B, D and E. Transverse sections. C and F. Longitudinal sections. Note that provascular bundles enclosed by yellow dashed lines became more distinguishable in (D-E) compared to (A-C). Green lines (a), (b), (d) and (e) represent the plane of transverse sections shown in (A), (B), (D) and (E), respectively. The detailed settings of frame average and number of projections are shown in Table S2. Scale bars = 1 mm. - The raw data will be saved as .raw files. Import the raw data into a built-in reconstruction software in the micro-CT system (e.g., coneCTexpress). Trim blank spaces, determine axes for coronal/sagittal sections, reduce noises using the noise reduction filter and convert the raw data into 8-bit Tiff images. Save the reconstructed Tiff image dataset.
Note: To compare multiple samples, a uniform setting in this step will be important. - Before importing the Tiff files into an image analysis software OsiriX, we rename them as following: Date_sample_resolution (µm)_number.tiff. This is important so that the image files are read by OsiriX in the correct order.
- Put a piece of clay on the sample stage in the micro-CT equipment and fix the sample tube tightly on the clay (Figure 4B).
- Constructing scanning movie from micro-CT data
Here, we introduce how to make a scanning movie using the software OsiriX to explore the micro-CT data (Video 1).Video 1. A 2D movie showing transverse sections of a maize stem. Green bars represent the plane shown in the transverse window. Structural annotations were added using Adobe Premiere Pro CC.- To import Tiff image data, disable ‘Import DICOM file format only’ option in the preferences of OsiriX. (Top Menu ‘OsiriX MD’ → ‘Preferences’ → ‘Database’ icon → ‘File Management window’ → uncheck the box of ‘Import DICOM file format only’.)
- Import Tiff images. (‘Import’ icon → Select the data folder. → ‘Copy Links’)
- To display the imported data, select the data file shown in ‘Documents DB window’ and click ‘2D Viewer’ icon. Then, select ‘3D Volume Rendering’ from the ‘3D Viewer’ in the top menu and enter resolution information in the fields of pixel X, Y and slice interval (Figure 10A).
Notes:- If OsiriX gives an error message, click ‘Continue’.
- The unit of the resolution is mm/pixel. Convert the resolution in µm/pixel into mm/pixel (e.g., 30 µm/pixel → 0.030 mm/pixel).
- If OsiriX gives an error message, click ‘Continue’.
- After clicking ‘OK’, a new window will open. Close this window.
- At this point, ‘Orientation’ icon has become active (Figure 10B). Now we can set the direction of 2D sections shown in the display using this icon and explore the sections using the slider located on the upper side of the 2D window.
- Open additional two windows by clicking ‘Window’ icon and selecting ‘3 window’ (Figure 10C). Enter the resolution information for these two windows as described in the Steps D3 and D4. Display coronal, sagittal and transverse sections in each window using ‘Orientation’ icon (Figures 10B and 9D).
- Select settings for window appearance using ‘WL/WW & CLUT’ and ‘Thick Slab’ functions. Our setting is below:
WL/WW & CLUT (Figure 10E (a))
WL/WW (window level/window width): Other
CLUT (color look up table): B/W Inverse
Opacity: Linear Table
Thick Slab (Figure 10E (b))
Mode: MIP
Slider: 5 - Adjust image WL/WW, position, magnification and angle in the window using tools shown in Figure 10E (c), (d), (e) and (f), respectively.
- To adjust size and arrangement of each window for the movie preparation, disable ‘Magnetic Windows for move & resize’. (Top Menu ‘OsiriX MD’ → ‘Preferences’ → ‘Viewers’ icon → uncheck ‘Magnetic Windows for move & resize’.) Adjust size and arrangement of windows (e.g., Figure 10F). Also, you can change the magnification of sections in each window using icon shown in Figure 10E (e).
- Export a movie file. (Select a window of transverse sections → ‘Movie Export’ icon → Set the data area exported as a movie and check the box of ‘Include all displayed 2D viewers’ (Figure 10G). Select the directory, set the format and frame rate. → Save the movie.
Note: As default, ‘Basic’ option is selected in ‘Annotations’ function area and various information about source data will be displayed in the 2D windows. You can disable this setting by checking ‘Graphic’.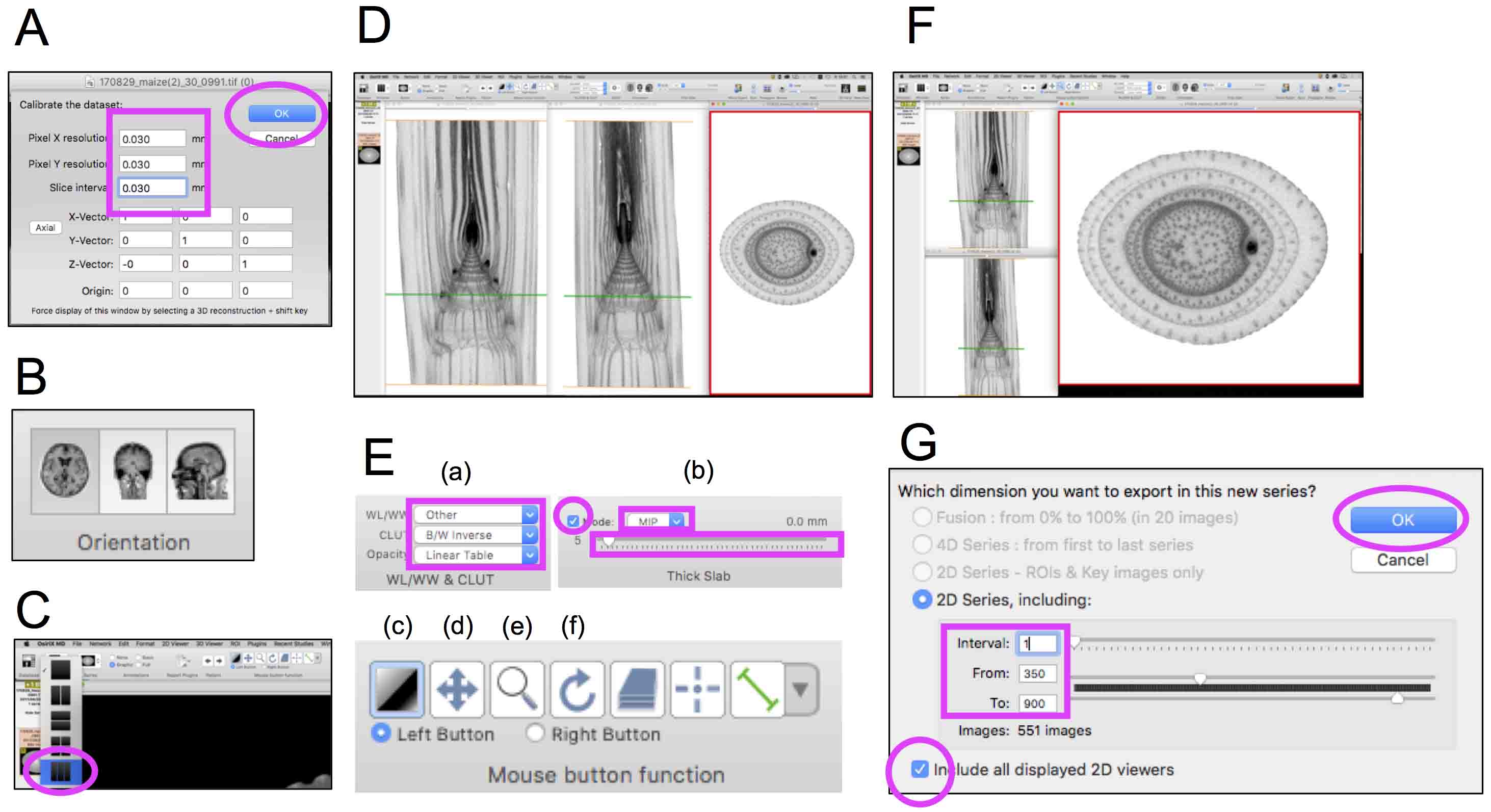
Figure 10. Movie preparation of the maize stem micro-CT data using OsiriX for easy observation. Magenta boxes and circles highlight icons, check boxes and numeric input fields selected in each step. A. A window to set the resolution; B. Orientation icons; C. Window icon to select ‘3 windows’; D. Three windows displaying different sections of the same sample. Left, middle and right windows display coronal, sagittal and transverse sections, respectively. E. Tools and icons used to modify window appearances. (a) WL/WW & CLUT, (b) Thick slab, (c) WL/WW, (d) position, (e) magnification and (f) angle. F. Arranged and resized windows to produce a movie. Note that the red-framed window displaying transverse sections is selected and active. Green bars in coronal and sagittal windows represent planes shown in transverse sections. G. A window to set parameters to export a movie.
- To import Tiff image data, disable ‘Import DICOM file format only’ option in the preferences of OsiriX. (Top Menu ‘OsiriX MD’ → ‘Preferences’ → ‘Database’ icon → ‘File Management window’ → uncheck the box of ‘Import DICOM file format only’.)
- Creating 3D model of maize stem vascular networks
Here, we extract vein traces and reconstruct vascular networks in maize stems using Imaris, a software which can be used in various analysis of 3D data including the extraction of filamentous structure such as veins. As this procedure includes many steps of manual operation, we provide videos to explain how to determine vein traces in 3D data (Videos 2 and 3).- Preparation of a 3D model of the maize stem in Imaris
- Import the sequential Tiff data into Imaris (Top Menu ‘Surpass’ icon → ‘File’ → ‘open’ → Select the first Tiff image in the data folder. → ‘Open’. Video 2: 0~10 sec).
- Enter the scale information (Top menu ‘Edit’ → ‘Image Properties’ → ‘Geometry’ (Figure 11A) → Enter the resolution (µm/pixel) → ‘OK’ → The 3D object often becomes very small after entering the resolution. Resize and fit the object to the window using ‘Fit’ icon at the bottom of the screen. Video 2: 12~27 sec).
- Change the model color. We selected white in this example. (Top menu ‘Edit’ → ‘Show Display Adjustment’ → ‘Channel 1’ (Figure 11B) → ‘Base color’ tab (Figure 11C) → Select color. Video 2: 33~38 sec).
- Change the model size and angle. (Video 2: 38~43 sec)
Note: You can rotate, move and enlarge/reduce the imported 3D model using left-dragging, right-dragging and wheeling actions of your computer mouse, respectively. - Crop 3D model to determine the data area analyzed. In this example, we cropped the stem into a quarter cylinder (‘Setting’ tab (Figure 11D) → ‘Blend’ → Top menu ‘Edit’ → ‘Crop 3D’ → Crop the 3D model (Figure 11E). → ‘OK’. Video 2: 44 sec~1 min 13 sec).
- Save the cropped 3D model. (Top menu ‘Export’ icon → Enter file name → ‘Save’. Video 2: 1 min 14 sec~1 min 35 sec).
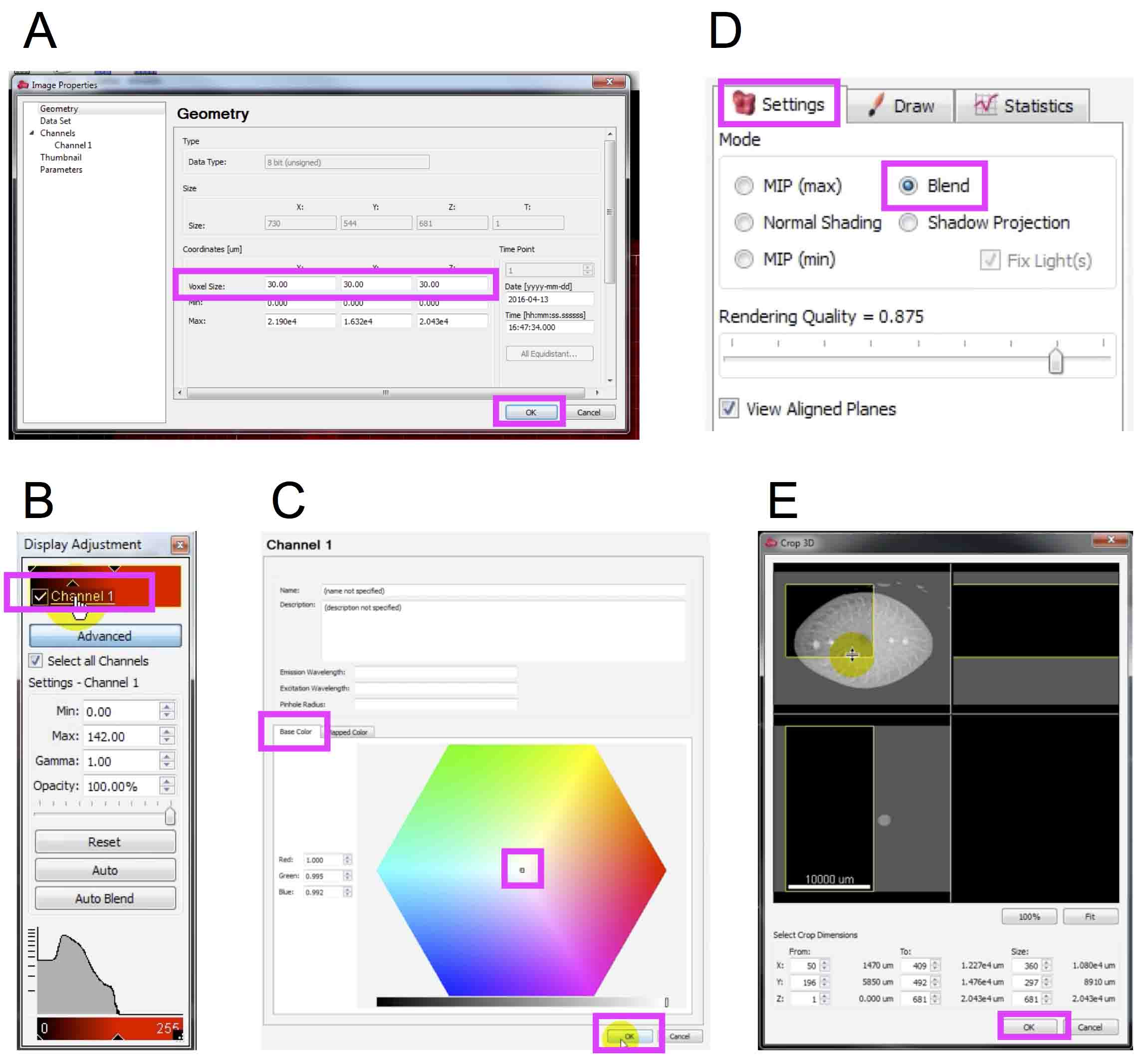
Figure 11. Preparation of a 3D model of the maize stem in Imaris. Magenta boxes highlight icons, check boxes and numeric input fields selected in each step. A. Geometry window to set resolution; B. Display adjustment window; C. Color selection window; D. Setting tab in the property area of the selected volume object; E. Crop 3D window.Video 2. A movie introducing procedures to prepare the maize stem 3D model in Imaris
- Import the sequential Tiff data into Imaris (Top Menu ‘Surpass’ icon → ‘File’ → ‘open’ → Select the first Tiff image in the data folder. → ‘Open’. Video 2: 0~10 sec).
- Tracing veins to create 3D model of vascular networks
Vein traces are the combination of relatively straight lines in internodes and winding lines in nodes. To make a vein trace from the micro-CT data, we divide a vein into a few segments depending on the position (internode or node), extract them independently and join them into a single trace.- Change settings of FilamentTracer to extract veins manually (Top menu ‘3D view’ icon → ‘Filament’ icon (Figure 12A) → Click ‘Skip automatic creation, edit manually’ (Figure 12B) → ‘Draw’ tab → Check ‘Manual’, ‘Automatic Placement’ and ‘XZ Plane’ (magenta boxes in Figure 12C). Video 3: 0~14 sec).
Note: Setting ‘XZ Plane’ is to extract a vertical segment of a vein in internodes in the Step E2d. - As default, the volume object shown as ‘Volume’ in the object list area is selected. To display slices of the XZ plane, uncheck ‘Volume’, otherwise the slices hide behind the volume object. Determine a vein to be traced using the slider (the bottom magenta box in Figure 12C and Video 3: 15~24 sec).
- Before starting to trace a vein, check ‘Select’ in ‘Camera/Labels’ window and then adjust the line width to that of the vein by wheeling the computer mouse. The size of square around the mouse pointer represents the line width. (The magenta box in Figure 12D and Video 3: 26~31 sec).
Note: This step is important for Imaris to recognize a vein efficiently based on the brightness of the 3D data. - Manually trace a segment of the vein in an internode. (Check ‘Select’ in ‘Camera/Labels’ window (the magenta box in Figure 12D) → Trace the vein by left-dragging while pressing ‘Shift’ key. Video 3: 32~43 sec).
- Edit the width and color of the traced line (Line width: ‘Filament’ tab → ‘cylinder’ → Enter the width (Figure 12E, e.g., 150). Line color: ‘Color’ tab → ‘Base’ → Change the color (Figure 12F). Video 3: 44~55 sec).
- Correct the traced line based on the brightness of the 3D data (‘Edit’ tab → ‘Segment’ → Click and select the traced line. (The line color will be yellow.) → ‘Center’ (Magenta boxes in Figure 12G). Video 3: 56 sec~1 min 04 sec).
- Manually trace a segment of the vein running horizontally in the node (‘Draw’ tab → Check ‘Manual’ and ‘Automatic Placement’ and ‘XY Plane’ (a blue box in Figure 12C) → Confirm the start and end of the segment of the vein in the node using a slider (Figure 12C and 12H). → Trace the segment and edit the line as described in Steps E2c-E2f. Video 3: 1 min 05 sec ~1 min 55 sec).
- Manually trace a segment of the vein in the lower internode as described in Steps E2c-E2f (Video 3: 1 min 58 sec~2 min 18 sec).
- Join the upper two segments (‘Camela/Labels’ window → Check ‘Navigate’ (A blue box in Figure 12D) → Enlarge the joint between upper two segments → Check ‘Select’ in ‘Camera/Labels’ window (a magenta box in Figure 12D). → ‘Edit’ tab (blue boxes in Figure 12G) → ‘Point’ → While pressing ‘Ctrl’ key, click the lower end of the upper segment and the upper end of the lower segment. Then both ends turn into yellow. → ‘Join’ → Click a blue joint point. → ‘Delete’ → Click newly formed ends while pressing ‘Ctrl’ key. → ‘Join’. Video 3: 2 min 19 sec~2 min 42 sec).
- Join the lower two segments as described in the Step E2i. (Video 3: 2 min 42 sec~3 min 00 sec).
- Correct the whole vein trace based on the brightness of the 3D data as described in the Step E2f (Video 3: 3 min 05 sec~3 min 14 sec).
- Repeat Steps E2c-E2k to extract traces of other veins. An example of the extracted vein network is shown in Figure 12I.
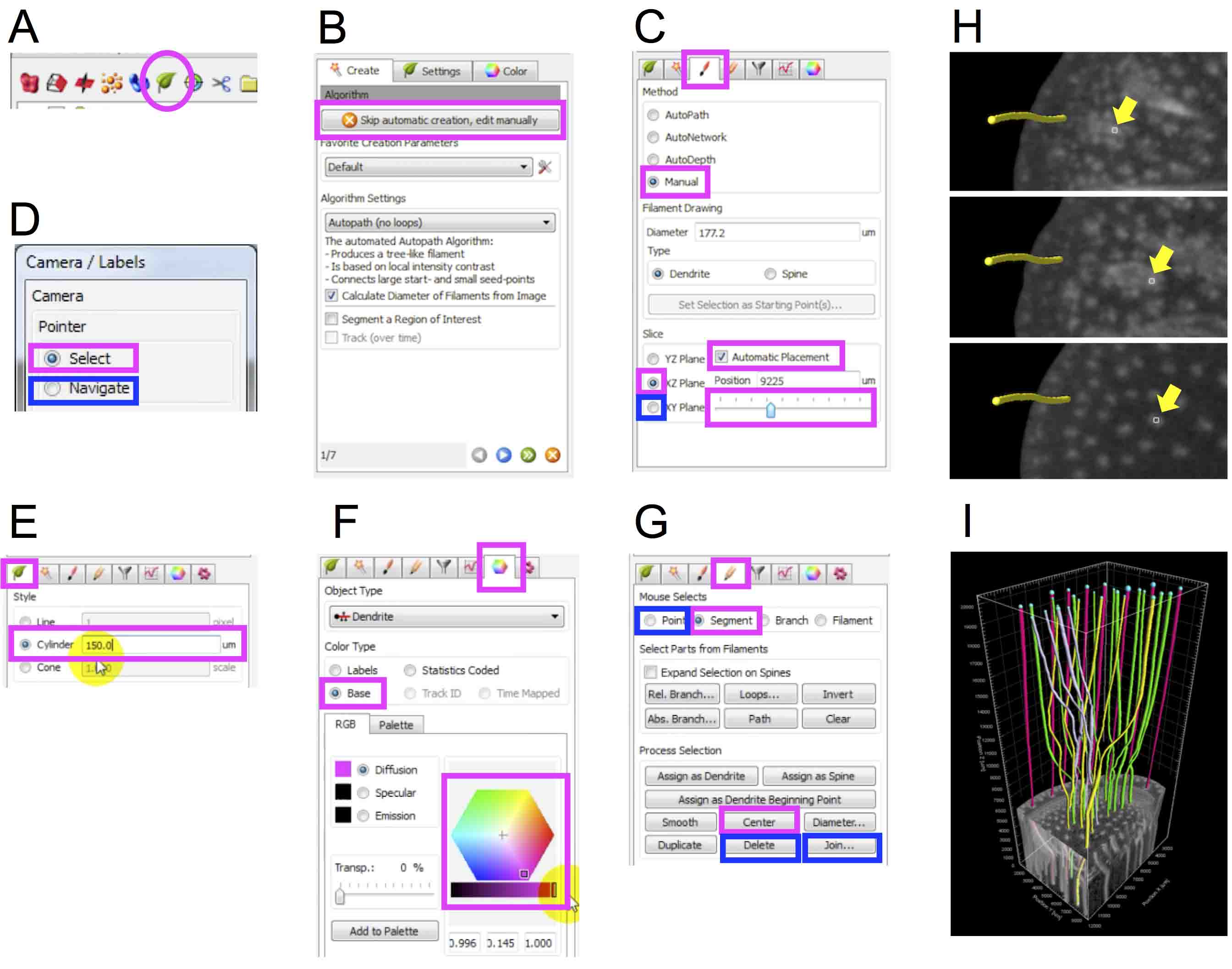
Figure 12. Tracing veins to create 3D model of vascular networks. Magenta boxes and circles highlight icons, check boxes and numeric input fields selected in each step. Blue boxes indicate those used in later steps. A. Filament icon; B. A button to skip automatic creation in the creation wizard window; C. Draw tab; D. Camera/Labels window; E. Setting tab; F. Color tab; G. Edit tab; H. Confirming the start and end of the segment running horizontally in the node. Top, middle and bottom panels represent the start, middle and end of the segment, respectively. Arrows indicate the vein to be traced shown in the node slices. I. A vascular network extracted from the micro-CT data of the maize stem.Video 3. A movie introducing procedures to extract vascular networks in the maize stem
- Change settings of FilamentTracer to extract veins manually (Top menu ‘3D view’ icon → ‘Filament’ icon (Figure 12A) → Click ‘Skip automatic creation, edit manually’ (Figure 12B) → ‘Draw’ tab → Check ‘Manual’, ‘Automatic Placement’ and ‘XZ Plane’ (magenta boxes in Figure 12C). Video 3: 0~14 sec).
- Preparation of a 3D model of the maize stem in Imaris
Data analysis
The raw data of micro-CT scanning was saved as .raw files and converted to 8-bit Tiff images using coneCTexpress as described in section C9. The 3D model of micro-CT scanning data was reconstructed from these tiff images using OsiriX (see Procedure D). The vascular network information was extracted using Imaris as described in Procedure E.
In our original research, we observed three independent biological replicates. Student’s t-test was applied for statistical analysis (Tsuda et al., 2017).
Recipes
- Fixative FAA solution
Formalin:acetic acid:50% ethanol = 5:5:90 - Iodine staining (Degenhardt et al., 2010)
Note: Tips for iodine staining using Lugol solution are well described by Gignac et al. (2016).
100% Lugol stock solution
Potassium iodide 10 g
Iodine 5 g
Distilled water (DW) to 100 ml - 25% Lugol working solution
Dilute 100% stock solution to 25% with DW upon use
Acknowledgments
We thank Mitsuhiko Kurusu (National Institute of Genetics, Office for Research Development) for organizing imaging seminars which led us to collaborate in this project and Akatsuki Kimura (National Institute of Genetics, Cell Architecture Laboratory) for introducing and maintaining Imaris. We also thank Sarah Hake (University of California, Berkeley) for providing maize stem samples. This work was supported by JSPS KAKENHI Grant Number 17H00440 to A.M. and JP16K18637 to K.T. This protocol was adapted from our recent work (Tsuda et al., 2017). The authors declare no conflicts of interest or competing interest.
References
- Degenhardt, K., Wright, A. C., Horng, D., Padmanabhan, A. and Epstein, J. A. (2010). Rapid 3D phenotyping of cardiovascular development in mouse embryos by micro-CT with iodine staining. Circ Cardiovasc Imaging 3(3): 314-322.
- Gignac, P. M., Kley, N. J., Clarke, J. A., Colbert, M. W., Morhardt, A. C., Cerio, D., Cost, I. N., Cox, P. G., Daza, J. D., Early, C. M., Echols, M. S., Henkelman, R. M., Herdina, A. N., Holliday, C. M., Li, Z., Mahlow, K., Merchant, S., Muller, J., Orsbon, C. P., Paluh, D. J., Thies, M. L., Tsai, H. P. and Witmer, L. M. (2016). Diffusible iodine-based contrast-enhanced computed tomography (diceCT): an emerging tool for rapid, high-resolution, 3-D imaging of metazoan soft tissues. J Anat 228(6): 889-909.
- Metscher, B. D. (2009a). MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol 9: 11.
- Metscher, B. D. (2009b). MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn 238(3): 632-640.
- Staedler, Y. M., Masson, D. and Schonenberger, J. (2013). Plant tissues in 3D via X-ray tomography: simple contrasting methods allow high resolution imaging. PLoS One 8(9): e75295.
- Tsuda, K., Abraham-Juarez, M. J., Maeno, A., Dong, Z., Aromdee, D., Meeley, R., Shiroishi, T., Nonomura, K. I. and Hake, S. (2017). KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell 29(5): 1105-1118.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Maeno, A. and Tsuda, K. (2018). Micro-computed Tomography to Visualize Vascular Networks in Maize Stems. Bio-protocol 8(1): e2682. DOI: 10.21769/BioProtoc.2682.
Category
Plant Science > Plant developmental biology > General
Cell Biology > Tissue analysis > Tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












