- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Behavioral Assays to Study Oxygen and Carbon Dioxide Sensing in Caenorhabditis elegans
Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2679 Views: 6927
Reviewed by: Khyati Hitesh ShahAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Multiplexing Thermotaxis Behavior Measurement in Caenorhabditis elegans
Stephan Raiders [...] Aakanksha Singhvi
Apr 5, 2022 2755 Views

Pharyngeal Pumping Assay for Quantifying Feeding Behavior in Caenorhabditis elegans
Muniesh Muthaiyan Shanmugam and Pankaj Kapahi
Sep 20, 2024 1769 Views

Station Holding During Rheotaxis: A Sensitive Assay of Lateral Line Function in Larval Zebrafish
Sophie Cohen-Bodénès [...] Lavinia Sheets
Dec 20, 2025 762 Views
Abstract
Animals use behavioral strategies to seek optimal environments. Population behavioral assays provide a robust means to determine the effect of genetic perturbations on the ability of animals to sense and respond to changes in the environment. Here, we describe a C. elegans population behavioral assay used to measure locomotory responses to changes in environmental oxygen (O2) and carbon dioxide (CO2) concentrations. These behavioral assays are high-throughput and enable examination of genetic, neuronal and circuit function.
Keywords: C. elegansBackground
Oxygen concentration provides C. elegans with information regarding environmental conditions. In laboratory conditions, when presented with an O2 gradient, C. elegans migrate towards intermediate concentrations (2%-12%) (Gray et al., 2004). Low levels of O2 may indicate the presence of bacteria (food) while high O2 levels may imply that the worms are close to the surface of its environmental substrate. Therefore, C. elegans responds in an exquisitely sensitive manner to changes in O2 concentration to enable navigation to optimal environments conducive to survival and propagation of offspring (Gray et al., 2004; Chang et al., 2006; Zimmer et al., 2009). Similarly, worms present a strong behavioral response to changes in CO2. Well-fed animals avoid CO2 while starved animals are attracted to CO2 (Hallem and Sternberg, 2008). This change in response may provide an evolutionary advantage to find food, as the bacterial food source releases CO2. Furthermore, pathogens generate CO2, which possibly indicates why well-fed worms avoid CO2. Specific neurons regulate gas sensing responses in C. elegans including the head neurons URXL/R, BAGL/R, AQR and the PQR neuron located in the tail (Hallem and Sternberg, 2008; Zimmer et al., 2009; Bretscher et al., 2011). The main regulators of O2 sensing are the URX and BAG neurons, which sense upshifts and downshifts of oxygen respectively. Regarding changes in CO2 levels, the BAG neurons are the principal sensors.
Materials and Reagents
- Worm pick made with platinum wire (Tritech Research, catalog number: PT-9901 )
- Filter paper (110 mm diameter) (GE Healthcare, Whatman, catalog number: 1001-110 )
- Petri dish 140 mm diameter (VWR, catalog number: 391-1500 )
- 90 mm Petri dishes (Techno Plas, catalog number: S9014UV20 )
- C. elegans strains
The following protocol applies to strains derived from wild type animals (N2, Bristol strain). Suggested controls (available from the Caenorhabditis Genetics Center (CGC)):- N2 (wild-type): positive control
- gcy-31(ok296): unable to respond to downshifts of O2 levels
- gcy-35(ok769): unable to respond to upshifts of O2 levels
- gcy-9(n4470): unable to respond to changes in CO2 concentration
- tax-4(p678): deficient in O2 and CO2 sensing
- N2 (wild-type): positive control
- Copper(II) chloride dihydrate (CuCl2·2H2O) (Sigma-Aldrich, catalog number: 221783-100G )
- NGM maintenance plates seeded with OP50 E. coli (see Recipes)
- KPO4 buffer (1 M, see Recipes)
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P0662-500G )
- Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: 3786-500G )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P0662-500G )
- Assay NGM plates (see Recipes)
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653-250G )
- Agar (SERVA high-gel strength agar) (SERVA Electrophoresis, catalog number: 11396.03 )
- Cholesterol (Sigma-Aldrich, catalog number: C8667-25G )
- Calcium chloride (CaCl2) (Merck, catalog number: 1.02382.0500 )
- KPO4 buffer (1 M)
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653-250G )
- Starvation plates (see Recipes)
Equipment
- Computer compatible with the software
Intel core i7, 32GB RAM, Windows 7, 64 bit. Needs one additional gigabit Ethernet card for connecting the GigE camera (Figure 1) - Camera
We use a 4-megapixel CCD camera (JAI, model: BM-500GE ), 35 mm objective: QIOPTIQ MEVIS 3516. Extension rings: PENT EXTENSION RING 1 (1 mm C-Mount) (Figure 1)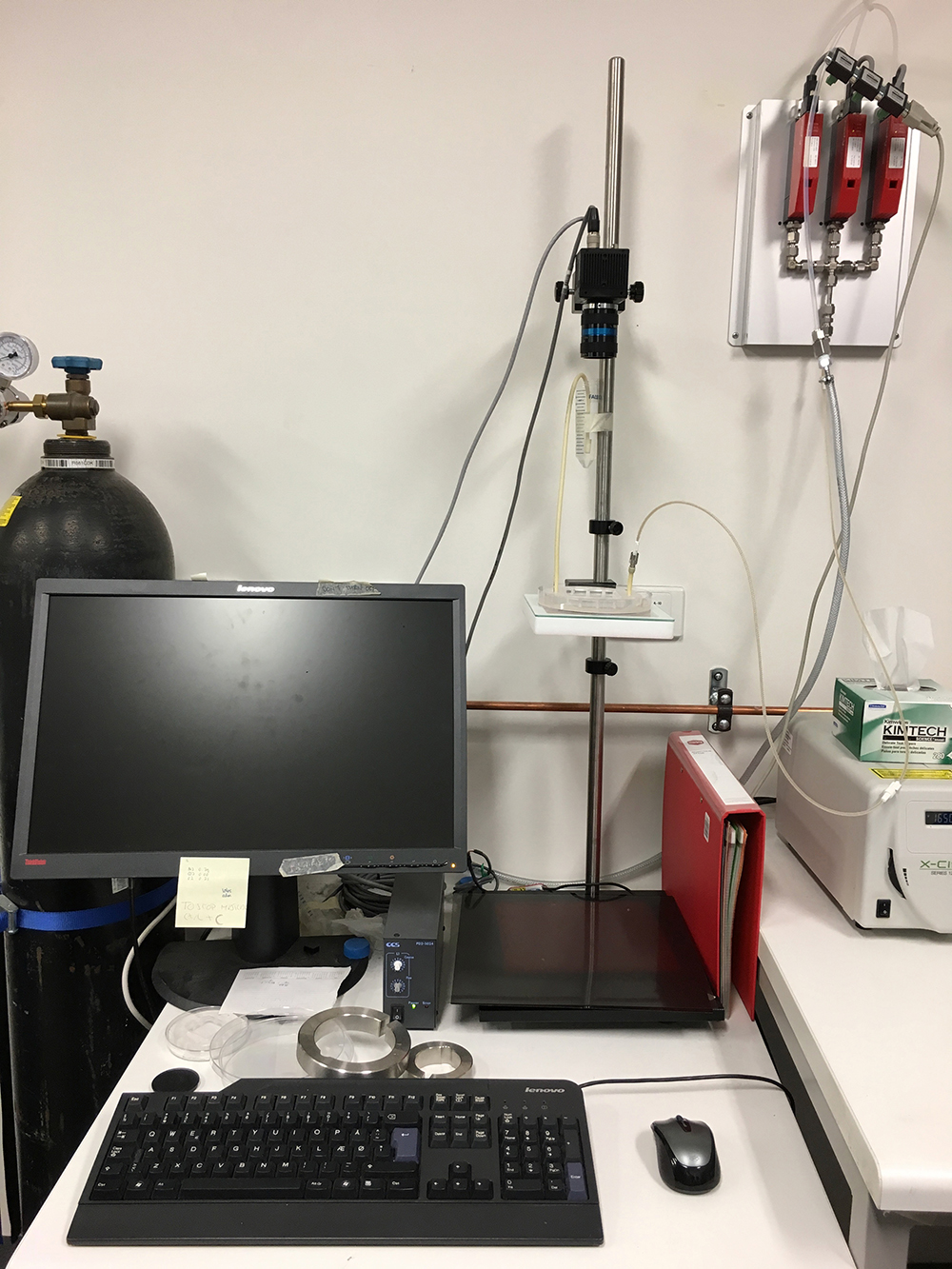
Figure 1. Computer and camera setup - Flow controllers
Vögtlin Instruments, CO2: GSC-A9TA-BB21 50 mln/min, N2: GSC-A9TA-BB22 200 mln/min, O2: GSC-A9TA-BB21 50 mln/min (Figure 2)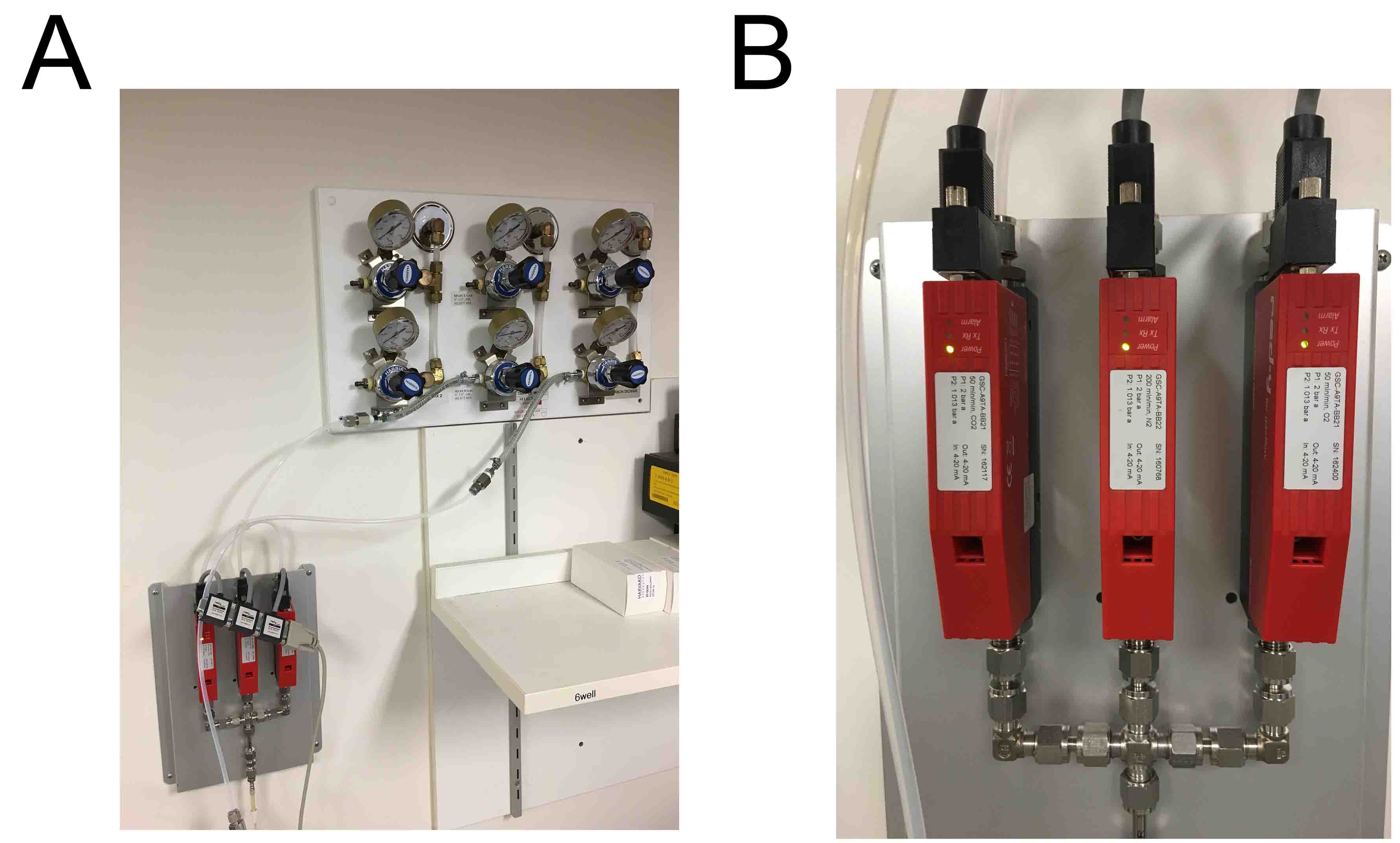
Figure 2. Gas and flow controller setup. A. Overview of mains gas supply connected to the flow controller. B. Close-up of the flow controller. - LED illumination (CCS, catalog number: TH-211/200RD ), cable (CCS, catalog number: FCB-1 ), power supply (CCS, catalog number: PD2-5024 )
- Individual gas bottles for O2, CO2 and N2
Note: Alternatively, premixed bottles may be used, depending on the flow controllers that are used. Nevertheless, we recommend individual gas bottles as they provide flexibility on the gas parameters that can be tested. - Assay chamber: custom-made transparent Plexiglas device with a flow arena of 60 x 60 x 0.7 mm (Figure 3)
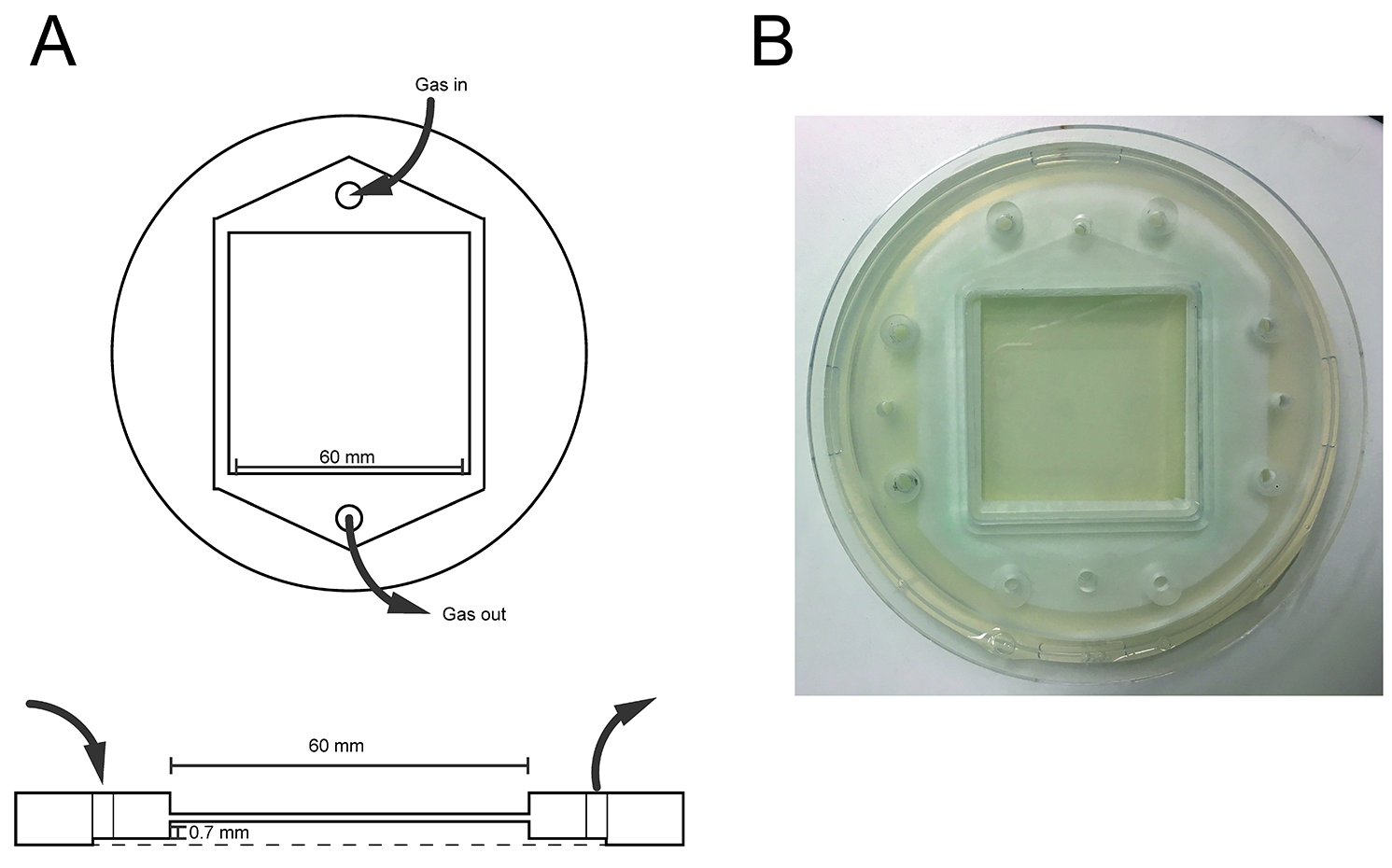
Figure 3. Custom-designed assay chamber. A. Top diagram: view from the top. Bottom diagram: lateral view. Note that the diagram is not scaled. B. Photograph of the assay chamber taken from above. - Autoclave
- Dryer (Binder, catalog number: 9010-0102 )
- 500 ml autoclaved bottle
Software
- Labview (to program and regulate the flow controllers)
- MATLAB (R13) (MathWorks) and Image Acquisition and Image Processing Toolbox (Figure 4)
- Parallel WormTracker (freely available: https://sourceforge.net/projects/wormtracker/)
- Streampix software (Norpix)
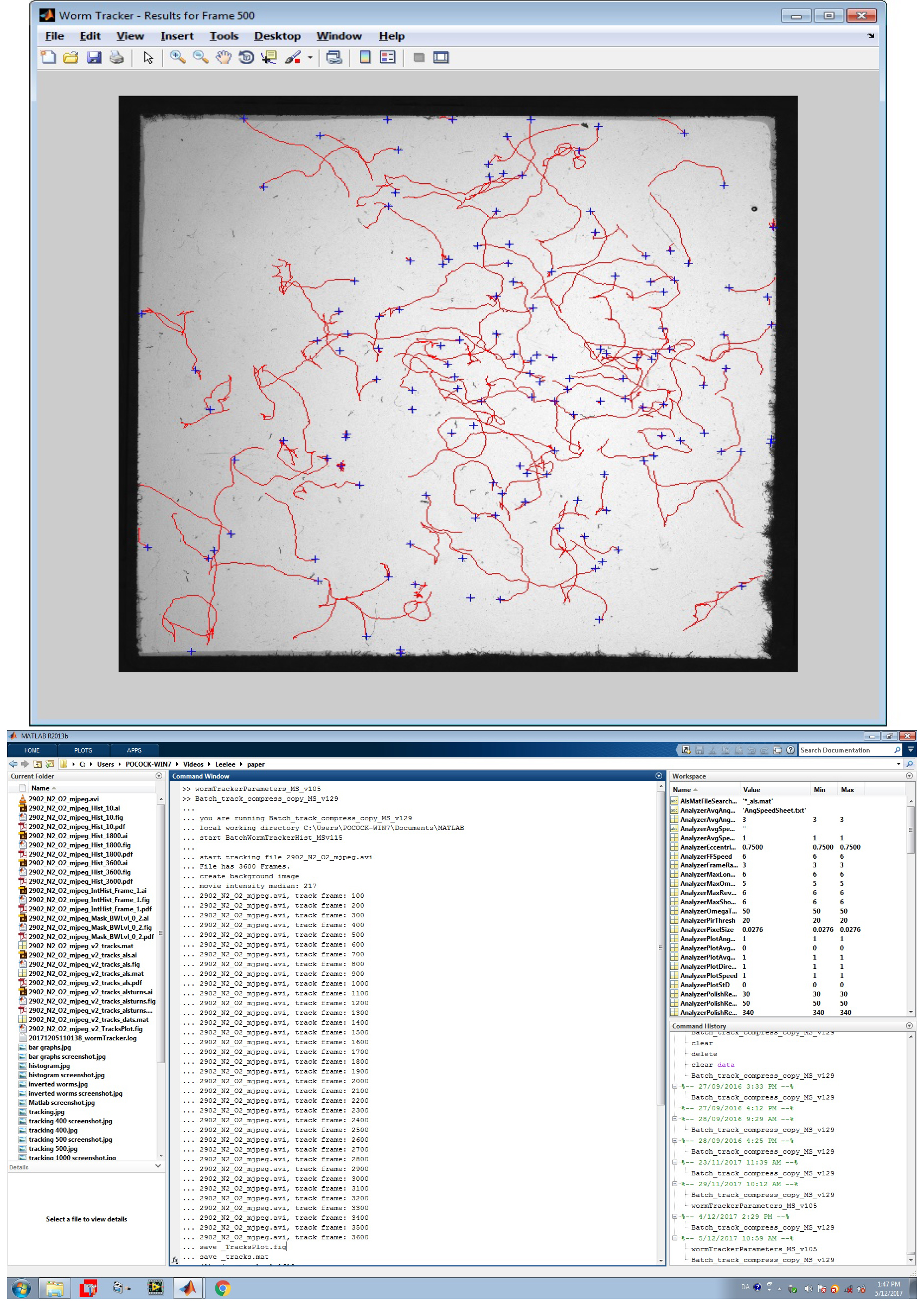
Figure 4. Screenshots of WormTracker analysis. A. Tracks of individual worms (red lines) detected by the software. B. MATLAB analysis software screen showing tracking frames.
Procedure
- Growth and synchronization of C. elegans populations
Recommendation: For each strain, pick 4 larval stage 4 (L4) hermaphrodites onto 4 maintenance plates every day (4 worms per plate), and use the following generation in the assay: For transgenic extrachromosomal strains, more than 4 transgenic L4s should be picked to ensure enough transgenic worms are available for the assay. In addition, for mutants with a lower brood size than wild type, a higher number of worms must be picked for synchronization. - Pick approximately 150 L4/young adult worms to a starvation plate (not seeded with OP50 E. coli). So that bacteria are not transferred to the empty plate, do not pick using bacteria, instead use the worm pick as a spoon to transfer worms that have crawled off the bacterial lawn.
Note: Worms should not be prepared by washing off a plate with M9 buffer, as behavioural assays are very sensitive to stress. - Allow worms to starve for 1 h. Feeding status strongly affects behavioral responses to O2 and CO2, therefore, the starvation must be conducted for the same length of time for each strain tested. Furthermore, well-fed N2 worms do not respond to downshifts in O2 (Zimmer et al., 2009).
- Prepare the assay plate (Figure 5)
- Cut a 56 x 56 mm squared area (a hole puncher may be used) in the center of the Whatman filter paper.
- Place the filter paper on top of the behavioral assay plate (14 cm NGM assay plate).
- Soak the filter paper at the border of the square with 20 mM CuCl2 to corral worms within the assay area.
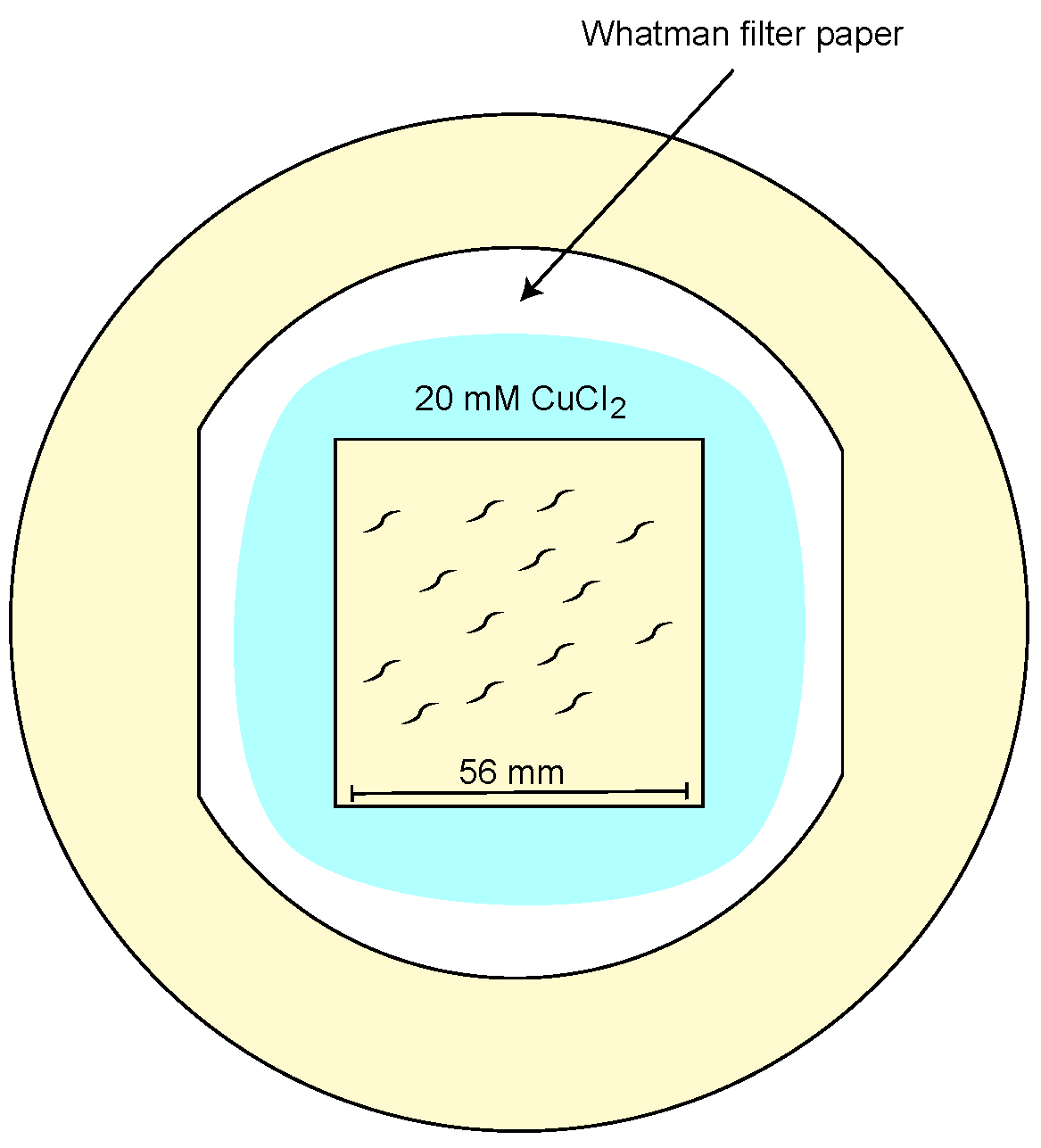
Figure 5. Illustration of the assay plate. 14 cm NGM assay plate containing a 56 x 56 mm arena of Whatman filter paper soaked in 700 μl of 20 mM CuCl2. Hermaphrodites are placed in the arena and the assay chamber is carefully positioned on top of the plate.
- Cut a 56 x 56 mm squared area (a hole puncher may be used) in the center of the Whatman filter paper.
- Spoon the worms from the starvation plate to the centre of the assay plate.
Note: Be careful not to damage the agar as worms will burrow. - Place the assay chamber on top of the assay plate. It is important that the 56 x 56 mm square within which the worms are located fits within the square of the chamber.
- Connect gas tubes to the chamber and start the gas flow with atmospheric concentrations (or alternatively with your desired starting conditions). Allow the gas to flow for at least 5 min prior commencing your experiments.
- Record videos while running the chosen gas concentration test.
Note: To assay O2 and CO2 sensing, balance the concentrations with N2. For testing acute O2 sensing, we suggest the following timings for each condition: 6 min 21%O2/79%N2–6 min 10%O2/90%N2–6 min 21%O2/79%N2. For testing acute CO2 sensing, we suggest the following timings for each condition: 6 min 21%O2/79%N2–6 min 21%O2/78%N2/1%CO2–6 min 21%O2 79%N2.
Data analysis
The parallel WormTracker software package includes a detailed user manual that describes how to run it for extraction of necessary data from the videos (Chalasani et al., 2007; Ramot et al., 2008). It is important to state that the script can be updated to different operating systems and video formats. The original script was written for PC running Windows XP and uncompressed, grayscale (8-bit) movies in AVI format with a resolution of 640 x 480. The WormTracker identifies worms and tracks their position defined by the worm’s centre of mass. The tracking is performed after the video has been recorded. After the worms are tracked, the Wormanalyzer permits analysis of the tracks and detection of the speed of the worms and other parameters such as turning events. Those data can be extracted and further analysed with any standard statistical software such as GraphPad Prism.
Representative data
When wild type hermaphrodites experience changes in O2 levels, they reduce their speed and change direction. Using the population behavioral assay, we describe in this protocol how these changes of speed are measurable to enable evaluation of the gas-sensing capability of a population of worms. We have previously used these assays to determine the functional importance of multiple transcription factors required for the development of gas-sensing neurons in C. elegans (Brandt et al., 2012; Gramstrup Petersen et al., 2013; Rojo Romanos et al., 2015 and 2017). In Figure 6, we illustrate examples of results obtained: we show how wild type worms decrease their speed in response to a BAG neuron-mediated downshift (21%-10%) or a URX neuron-mediated upshift (10%-21%) in O2 concentration (Figure 6A). In contrast, lin-32(tm1446) mutant animals, which lack the LIN-32/Atoh1 transcription factor and have defects in URX development, fail to respond to an upshift in O2 concentration from 10% to 21% (Figure 6B) (Rojo Romanos et al., 2017). Finally, egl-13(ku194) mutant animals, in which the EGL-13/Sox transcription factor is deleted, fail to respond to upshifts and downshifts in O2 concentration due to a defect in specification of the BAG and URX neurons (Figure 6C) (Gramstrup Petersen et al., 2013).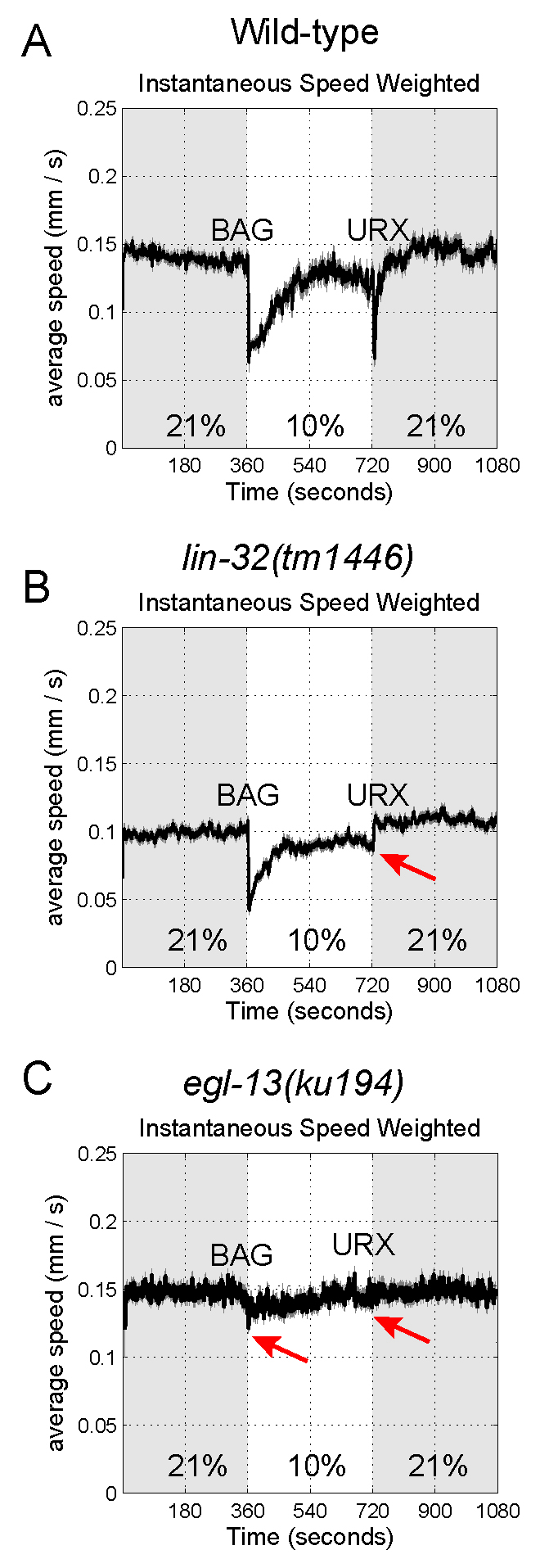
Figure 6. C. elegans mutants that fail to respond to changes in O2 levels. Graphs showing the locomotion speed of wild-type (A), lin-32(tm1446) (B) and egl-13(ku194) (C) mutant animals when O2 levels are shifted every 6 min: 21%-10%-21%. The data represent averages of at least four independent assays (80-120 animals per assay). lin-32(tm1446) mutants fail to respond to O2 upshifts (URX-mediated) but show a similar response to wild type animals to O2 downshifts (BAG-mediated). In contrast, egl-13(ku194) mutant animals fail to respond to O2 upshifts and downshifts due to defects in BAG and URX specification. Red arrows indicate defective responses to changes in O2 levels.
Recipes
- Maintenance plates
See He (2011) - KPO4 (1 M)
Prepare separate 1 M solutions of KH2PO4 (Sigma-Aldrich) and K2HPO4 (Sigma-Aldrich)
Filter sterilize both solutions and measure 434 ml of 1 M KH2PO4 and 66 ml of 1 M K2HPO4 using an autoclaved measuring cylinder before transferring to a 500 ml autoclaved bottle - Assay NGM plates (22 g/L agar)
Use 14 cm plates (VWR, Petri dish 140 mm diameter)- For 1 L NGM:
3 g NaCl
22 g agar (SERVA high-gel strength agar)
1 ml cholesterol (5 mg/ml) (add after autoclaving)
1 ml CaCl2 (1 M) (add after autoclaving)
25 ml KPO4 (1 M) (add after autoclaving) - Pour precisely 75 ml of NGM solution into each 14 cm plate and allow to solidify on a stable and perfectly flat surface
- When the agar is solid, remove residual water condensed on the lid of the plate with tissue paper
- Dry the plates in a dryer (Binder) overnight to 24 h at 50 °C with the lid facing down
- Take plates out of the dryer and remove residual water on the lid of the plate with tissue paper
- Leave plates at room temperature for one day
- Use these plates directly for behavioral experiments or store them in the cold room/fridge for up to 30 days
- For 1 L NGM:
- Starvation plates (17 g/L agar)
Use 9 cm plates (90 mm Petri dishes, Techno Plas)
For starvation plates use the same recipe as for assay plates but with 17 g agar per liter (instead of 22 g per liter)
Note: Do not seed these plates with bacteria.
Acknowledgments
Some strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and by Shohei Mitani at the National Bioresource Project (Japan). This work was supported by a grant from the European Research Council (ERC Starting Grant number 260807), Monash University Biomedicine Discovery Fellowship and veski innovation fellowship: VIF 23 to R.P. This protocol has been adapted from (Zimmer et al., 2009; Rojo Romanos et al., 2017).
The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC (grant agreement 281869) and the Research Institute of Molecular Pathology (IMP). The IMP is funded by Boehringer Ingelheim.
Conflict of interest statement: The authors declare no conflict of interest or competing interests.
References
- Brandt, J. P., Aziz-Zaman, S., Juozaityte, V., Martinez-Velazquez, L. A., Petersen, J. G., Pocock, R. and Ringstad, N. (2012). A single gene target of an ETS-family transcription factor determines neuronal CO2-chemosensitivity. PLoS One 7(3): e34014.
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77(1): 71-94.
- Bretscher, A. J., Kodama-Namba, E., Busch, K. E., Murphy, R. J., Soltesz, Z., Laurent, P. and de Bono, M. (2011). Temperature, oxygen, and salt-sensing neurons in C. elegans are carbon dioxide sensors that control avoidance behavior. Neuron 69(6): 1099-1113.
- Chalasani, S. H., Chronis, N., Tsunozaki, M., Gray, J. M., Ramot, D., Goodman, M. B. and Bargmann, C. I. (2007). Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 450(7166): 63-70.
- Chang, A. J., Chronis, N., Karow, D. S., Marletta, M. A. and Bargmann, C. I. (2006). A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol 4(9): e274.
- Gramstrup Petersen, J., Rojo Romanos, T., Juozaityte, V., Redo Riveiro, A., Hums, I., Traunmuller, L., Zimmer, M. and Pocock, R. (2013). EGL-13/SoxD specifies distinct O2 and CO2 sensory neuron fates in Caenorhabditis elegans. PLoS Genet 9(5): e1003511.
- Gray, J. M., Karow, D. S., Lu, H., Chang, A. J., Chang, J. S., Ellis, R. E., Marletta, M. A. and Bargmann, C. I. (2004). Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430(6997): 317-322.
- Hallem, E. A. and Sternberg, P. W. (2008). Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105(23): 8038-8043.
- He, F. (2011). Common worm media and buffers. Bio Protoc Bio101: e55.
- Ramot, D., Johnson, B. E., Berry, T. L., Jr., Carnell, L. and Goodman, M. B. (2008). The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS One 3(5): e2208.
- Rojo Romanos, T., Petersen, J. G., Riveiro, A. R. and Pocock, R. (2015). A novel role for the zinc-finger transcription factor EGL-46 in the differentiation of gas-sensing neurons in Caenorhabditis elegans. Genetics 199(1): 157-163.
- Rojo Romanos, T., Pladevall-Morera, D., Langebeck-Jensen, K., Hansen, S., Ng, L. and Pocock, R. (2017). LIN-32/Atonal controls oxygen sensing neuron development in Caenorhabditis elegans. Sci Rep 7(1): 7294.
- Zimmer, M., Gray, J. M., Pokala, N., Chang, A. J., Karow, D. S., Marletta, M. A., Hudson, M. L., Morton, D. B., Chronis, N. and Bargmann, C. I. (2009). Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron 61(6): 865-879.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rojo Romanos, T., Ng, L., Zimmer, M. and Pocock, R. (2018). Behavioral Assays to Study Oxygen and Carbon Dioxide Sensing in Caenorhabditis elegans. Bio-protocol 8(1): e2679. DOI: 10.21769/BioProtoc.2679.
Category
Neuroscience > Behavioral neuroscience > Sensorimotor response
Neuroscience > Sensory and motor systems > Animal model
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








