- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determining Ribosome Translational Status by Ribo-ELISA
Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2670 Views: 7874
Reviewed by: Alessandro DidonnaSteven BoeynaemsAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Mitochondrial Biogenesis Assay after 5-day Treatment in PC-3 Cells
Yili Xu and Joy Y. Feng
Jan 20, 2015 12054 Views

A Highly Sensitive, Reproducible Assay for Determining 4-hydroxynonenal Protein Adducts in Biological Material
T. Blake Monroe and Ethan J. Anderson
Oct 5, 2019 3957 Views

An Optimised Indirect ELISA Protocol for Detection and Quantification of Anti-viral Antibodies in Human Plasma or Serum: A Case Study Using SARS-CoV-2
Claire Baine [...] Jennifer Serwanga
Dec 20, 2023 3637 Views
Abstract
The Ribo-ELISA was originally developed to elucidate the basis for the ribopuromycylation method (RPM)-based detection of ribosome bound nascent chains. The Ribo-ELISA enables characterization of the translational status of ribosomes, and can be applied to the discovery of super-ribosomal complexes with novel ribosome associated macromolecules that are isolated by physical fractionation in sucrose gradients or other methods.
Keywords: RibosomeBackground
Ribosomes are heterogeneous structures consisting of 40S and 60S subunits that are present in cells as monosomes and polysomes, when multiple ribosomes are bound to a single mRNA. Additionally, translating ribosomes can be associated with multiple molecular complexes that modulate translation. The ribosome ELISA (Enzyme-Linked ImmunoSorbent Assay) enables detection of translating ribosomes by in vitro puromycylation of ribosome associated nascent chains (David et al., 2012). This chemical reaction proceeds spontaneously upon adding puromycin to ribosomes with bound nascent chains. This method quantitates the number of nascent chains present per ribosome and can be used to determine the translational status of monosomes relative to polysomes, and identify ribosomes bound to other macromolecules that alter its sedimentation rate or migration in sizing columns.
Materials and Reagents
- Materials
- 175 cm2 flasks (Corning, catalog number: 431306 )
- Thinwall 13.2 ml polypropylene tube (Beckman Coulter, catalog number: 331372 )
- Parafilm (Bemis, catalog number: 701606 )
- Pipette tips (MµltiGuardTM Tips 1-100 µl, 1-200 µl, 100-1,000 µl) (Sorenson BioScience, catalog numbers: 36060 , 14220 , 14200 )
- 1.5 ml Eppendorf tubes (Sigma-Aldrich, catalog number: Z666505 )
Manufacturer: Eppendorf, catalog number: 022431081 . - 96-well Millipore Multiscreen HTS PVDF plates (Merck, catalog number: MSIPS4W10 )
- Paper towels
- 175 cm2 flasks (Corning, catalog number: 431306 )
- Cell line(s)
Though this procedure was originally designed for HeLa cells, it can be easily adapted for any other cell lines. The described procedure and associated picture (Figure 1) uses HeLa cells (ATCC, catalog number: CCL-2.1 ) - Reagents
- Trypsin-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25300054 )
- StartingBlockTM blocking buffer (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 37538 )
- Hydrogen chloride (HCl) 1.6%
- Potassium phosphate monobasic (KH2PO4)
- Sodium chloride (NaCl)
- Na2HPO4·7H2O (Life Technologies)
- Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 41966029 )
- Penicillin/streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 10378016 )
- Fetal bovine serum (FBS) (Eurobio, catalog number: CVFSVF0101 )
- Tris-HCl pH 7.5 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15567027 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M2670 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Sucrose (Sigma-Aldrich, catalog number: 84097 )
- NP-40 10% (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 28324 )
- Complete Mini EDTA-free protease inhibitor tablets (Sigma-Aldrich, Roche Dignostics, catalog number: 11836170001 )
- RNase Out (Life Technologies, catalog number: 100000840 or Thermo Fisher Scientific, InvitrogenTM, catalog number: 10777019 )
- DEPC treated water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 750023 )
- 16% paraformaldehyde (Electron Microscopy Sciences, catalog number: 15710 )
- Glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- 70% ethanol (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R40135 )
- Trypsin-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25300054 )
- Protein synthesis inhibitors
- Cycloheximide (CHX) (Sigma-Aldrich, catalog number: C7698 )
Stock solution (1,000x): 100 mg/ml (or 355 mM) in 50% ethanol
Store at -20 °C - Emetine dihydrochloride (Emetine) (Sigma-Aldrich, catalog number: E2375 )
Stock solution (1,000x): 25 mg/ml (or 45 mM) in 50% ethanol
Store at -20 °C - Puromycin (PMY) (Sigma-Aldrich, catalog number: P7255 )
Stock solution (1,000x): 50 mg/ml (or 91 mM) in 50% ethanol
Store at -20 °C
- Cycloheximide (CHX) (Sigma-Aldrich, catalog number: C7698 )
- Antibodies
- Primary antibodies
- Anti-puromycin mouse monoclonal antibodies: hybridoma clone 2A4 is available to the scientific community from the Developmental Studies Hybridoma Bank http://dshb.biology.uiowa.edu/PMY-2A4
- Anti-ribosomal P antibody: human polyclonal autoimmune antiserum (from lupus patients) which recognizes three proteins of the 60S ribosomal subunit, RPLP0, P1 and P2 (ImmunoVision, catalog number: HPO-0300 )
- Anti-puromycin mouse monoclonal antibodies: hybridoma clone 2A4 is available to the scientific community from the Developmental Studies Hybridoma Bank http://dshb.biology.uiowa.edu/PMY-2A4
- Secondary antibodies
- Rabbit anti-mouse HRP conjugate (Cappel) (MP Biomedical, catalog number: 55564 )
- Goat anti-human Ig-HRP (Jackson ImmunoResearch, catalog number: 109-035-003 )
- Rabbit anti-mouse HRP conjugate (Cappel) (MP Biomedical, catalog number: 55564 )
- Primary antibodies
- Solutions (see Recipes)
- Phosphate buffered saline (PBS)
- Growth medium
- Homogenization buffer
- Polysome buffer
- 15% sucrose solution
- 50% sucrose solution
- 3% PFA
- Phosphate buffered saline (PBS)
Equipment
- PIPETMAN 1-20 µl (Sartorius, model: Proline® Plus, catalog number: 728030 )
- PIPETMAN 20-200 µl (Sartorius, model: Proline® Plus, catalog number: 728060 )
- PIPETMAN 100-1,000 µl (Sartorius, model: Proline® Plus, catalog number: 728070 )
- Water bath (JULABO, model: TW20 )
- Class II laminar flow hood (FASTER, model: SafeFAST Elite )
- Cell Incubator (Heraeus, model: HERACell )
- Centrifuge (Eppendorf, model: 5415 R )
- Inverted microscope (Nikon Instruments, model: Eclipse TS100 )
- Microplate photometer (TECAN, model: Infinite M200 )
- Dounce homogenizer (DWK Life Sciences, Kimble, catalog number: 885300-0002 )
- Ceramic bead tubes Lysing matrix D (MP Biomedicals, catalog number: 116913050 )
- FastPrep-24 MP device (MP Biomedicals, catalog number: 116004500 )
- Ultracentrifuge (Beckman Coulter)
- Rotor (Beckman Coulter, model: SW 41 Ti )
- MultiScreenHTS Vacuum manifold (Merck, catalog number: MSVMHTS00 )
- Optional: Density gradient fractionator (Teledyne Isco)
Software
- GraphPad Prism software
Procedure
The following procedure describes the first application of Ribo-ELISA (David et al., 2012) which was used to detect puromycin association with ribosomes: at that time it was not clear whether puromycin binding was limited to translating ribosomes.
Either emetine or CHX can be used as elongation inhibitors to freeze polysome. Unlike CHX, emetine irreversibly inhibits translation, and need not be maintained throughout the procedure.
Day 1:
- Pre-warm growth medium (see Recipes), PBS and trypsin-EDTA in 37 °C water bath. Trypsinize two 175 cm2 flasks of cells at 70%-80% confluency. Remove the medium, wash with 10 ml of warm PBS, add 5 ml of trypsin-EDTA and leave for 5 min at 37 °C. Resuspend cells with 5 ml of growth medium.
- Seed two 175 cm2 flasks with 1 x 107 HeLa cells per flask (Corning) containing 15 ml of warm growth medium each. Incubate for 30 h to allow HeLa cells to attach and proliferate.
- Sucrose gradients (see Video 1). Prepare 15% and 50% sucrose solutions (see Recipes). Add 5 ml of 50% sucrose solution in 13.2 ml Polypropylene centrifuge tube (Beckman Coulter). Tilt the tube (almost 90°) and carefully add 5 ml of 15% sucrose solution on the top (add slowly the solution flowing along the tube wall and slowly put the tube back to vertical). Seal the tube with Parafilm and gently put the tube in horizontal position on the bench. Wait 1-2 h and put it back to the vertical. The 15%-50% sucrose gradient is ready. Store it in the fridge O.N.
Note: Other methods can be used to prepare gradients. A convenient alternative is the freeze-thawing method which entails successive freezing (-80 °C) of 2 ml layered sucrose solutions starting from the highest sucrose concentration (50%, 40%, 30%, 20%, 10% w/v sucrose solutions) (Bastide et al., 2017).Video 1. Sucrose gradients preparation. This video provides step by step guidelines for preparing 15% and 50% sucrose gradients.
Day 2:
- Examine HeLa cells under an inverted microscope to ensure right confluence (between 60%-70%).
- Incubate cells for 5 min with 10 ml of warm growth medium supplemented with 100 µg/ml CHX (or 20 µg/ml emetine) added directly to the medium.
- Remove the medium, wash twice with 10 ml of warm PBS. Add 5 ml of trypsin-EDTA and put the cells back in the incubator for 5 min, then resuspend cells with 5 ml of growth medium, centrifuge at 400 x g for 5 min and wash the pellet twice with 10 ml of ice-cold PBS (400 x g, 5 min, 4 °C) (both solutions supplemented with 100 µg/ml CHX, see Recipes).
Note: From this step RNase free tips should be used. - Pellet cells by centrifugation (400 x g, 5 min, 4 °C). Resuspend cells in 1 ml ice-cold homogenization buffer (see Recipes).
- Transfer the cell suspension to ceramic beads tubes (Lysing matrix D) on ice.
Note: Importantly, NP-40 in homogenization buffer is not essential when using ceramic beads to lyse cells: ribosomes are recovered with similar efficiency. Therefore, Ribosome ELISA can be performed in the absence of detergent. - ‘Vortex’ at RT for 20 sec (using the FastPrep-24 MP device).
Note: In lieu of ceramic beads, cells can be stroked 10 times in ice cold homogenization buffer using Dounce homogenizer embedded in ice. - Transfer supernatant to 1.5 ml Eppendorf tubes. Spin at 13,000 x g for 10 min at 4 °C.
- Carefully collect the supernatant.
Note: If the supernatant is not perfectly clear, recentrifuge. - Slowly layer 1 ml clear lysate along the tube wall on the top of the cold 15-50% sucrose gradient. Balance the rotor with another sample or a tube filled with water. Spin at 151,263 x g (35,000 rpm) (Beckman Coulter, rotor SW41.Ti) for 2.5 h at 4 °C.
- Fractionate (1 ml fractions) and measure absorbance continuously at 254 nm using a gradient system automate (Teledyne Isco). You will get 11 fractions.
Note: If you lack access to an automated gradient collector, fractions can be collected in the old-fashioned way; manually from the top. Use a PIPETMAN 100-1,000 µl with a 1 ml tip. Push the piston to the 1 ml max, and place the tip in contact with the meniscus. Carefully, collect 1 ml sucrose by releasing the pipette piston slowly. Always keep the tip in contact with the meniscus. Put each fraction in a clean 1.5 ml tube (11 fractions). - Add 100 µl of each fraction to wells of a 96-well Millipore Multiscreen HTS PVDF plate (11 wells, lane 1). Repeat this operation five times in following lanes to get technical replicates (3 wells per fraction) for two experimental conditions (anti-puromycin staining and anti-P protein staining). Load an identical plate as control (‘no puromycin labeling’). Incubate plates for 10 min on ice, and then vacuum the liquid through using the MultiScreenHTS Vacuum manifold.
- Add 200 µl/well of polysome buffer (see Recipes) and vacuum again.
- Puromycin labeling. Add 100 µl of PMY (91 µM) containing polysome buffer to the experimental plate and 100 µl of polysome buffer to the control plate. Incubate for 10 min on ice. In parallel, skip this step in order to generate the ‘no puromycin labeling’ control plate.
- Wash. Flick plate out into sink, briefly drain inverted on paper towels. Add 200 µl of polysome buffer, flick plate out again. Repeat this operation twice.
- Fixation. Add 100 µl of 3% paraformaldehyde (PFA) (see Recipes) inside the hood. Incubate for 10 min at room temperature (RT). Dispose of the PFA waste properly.
Note: We used this fixation step in the original protocol to mimic the RPM procedure on cells. However, this step is not necessary to detect puromycylated nascent chains. - Wash. Wash wells with 200 µl PBS 3 times.
- Membrane blocking. Incubate with 100 µl of StartingBlockTM blocking buffer for 30 min at RT. Vacuum.
Note: In the original procedure, StartingBlockTM gave very low background Ab binding. Using different Abs, other blocking buffers might be superior. - Primary antibody. Incubate for 1 h with 50 µl of either 2.5 µg/ml anti-PMY mAb (in StartingBlockTM) at RT or anti-Ribosomal P antibodies (1/5,000, in StartingBlockTM)
- Wash. Wash wells with 200 µl PBS 3 times.
- Secondary antibody. Incubate with 50 µl of secondary antibody HRP conjugate, rabbit anti-mouse for PMY staining and goat anti-human for ribosomal P staining (1/100 in StartingBlockTM) for 1 h at RT.
- Wash. Wash wells with 200 µl PBS 3 times.
- Signal Generation. Add 100 µl of TMB substrate (KPL) to each well. When some blue color becomes visible in the well, stop the reaction using 50 µl of 1.6% HCl. Transfer the liquid in each well to a plastic plate with a transparent bottom. Then, measure absorbance at 450 nm.
- Quantification. Determine the level of background staining by using the average of unlabeled wells from the control plate and subtract it from test values. In the illustrated example, data were graphed using GraphPad Prism software (Figure 1).
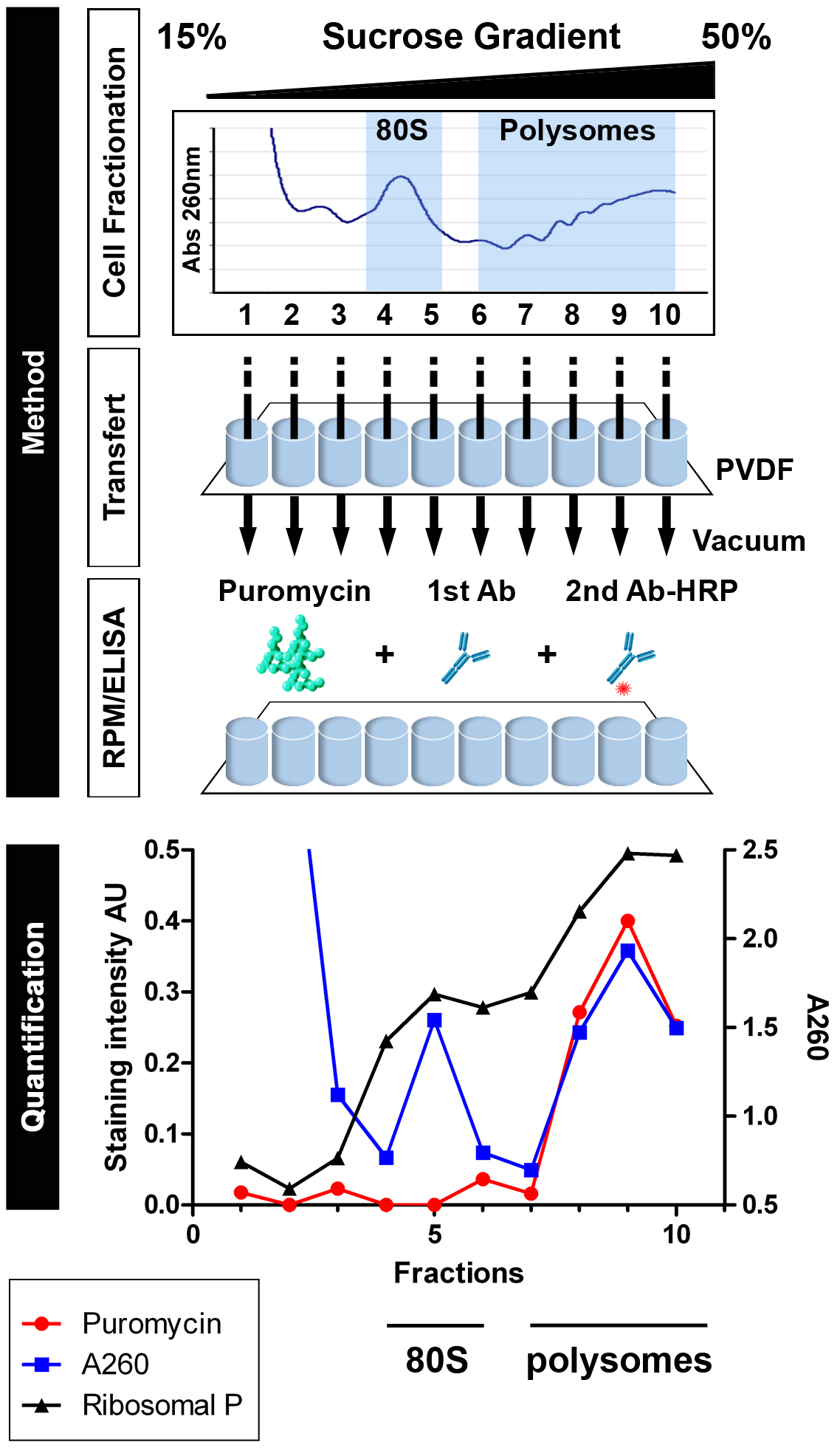 Figure 1. Schematic representation of Ribosome ELISA applied to detect PMY association with translating ribosomes (from David et al., 2012). HeLa cells or HeLa cells incubated with emetine for 15 min were lysed and fractionated on 15-50% sucrose gradients. Fractions were bound to PVDF 96-well plates and incubated with PMY, which results in ribosome-catalyzed puromycylation. Ribosomes were detected by A260 of fractions or by ELISA for the ribosomal P proteins (here resolved into the 3 known species) as detected by human autoimmune Abs, which establishes that 60S subunit and monosome (80S) were bound to PVDF (fractions 4 and 5). Puromycylation was detected by ELISA for PMY using 12-D10, and clearly demonstrates that monosomes and free 60S subunits do not stably associate with PMY, which requires nascent chains.
Figure 1. Schematic representation of Ribosome ELISA applied to detect PMY association with translating ribosomes (from David et al., 2012). HeLa cells or HeLa cells incubated with emetine for 15 min were lysed and fractionated on 15-50% sucrose gradients. Fractions were bound to PVDF 96-well plates and incubated with PMY, which results in ribosome-catalyzed puromycylation. Ribosomes were detected by A260 of fractions or by ELISA for the ribosomal P proteins (here resolved into the 3 known species) as detected by human autoimmune Abs, which establishes that 60S subunit and monosome (80S) were bound to PVDF (fractions 4 and 5). Puromycylation was detected by ELISA for PMY using 12-D10, and clearly demonstrates that monosomes and free 60S subunits do not stably associate with PMY, which requires nascent chains.
Data analysis
For data analysis, we previously used GraphPad Prism software. An example is presented in the following paper: David et al., 2012.
Notes
Polysome profile can vary dramatically depending on the cell line and translation rate.
Recipes
- Phosphate buffered saline (PBS)
210.0 mg/L KH2PO4
9,000 mg/L NaCl
726.0 mg/L Na2HPO4·7H2O (Life Technologies) - Growth medium
Dulbecco’s modified Eagle’s medium with glutamine (Thermo Fisher Scientific)
1% penicillin/streptomycin
7.5% fetal bovine serum (FBS) - Homogenization buffer
50 mM Tris-HCl pH 7.5
5 mM MgCl2
25 mM KCl
0.2 M sucrose (Sigma-Aldrich)
0.5% NP-40 (Thermo Fisher Scientific)
100 µg/ml CHX (Sigma-Aldrich)
EDTA-free protease inhibitors (Roche)
10 U/ml RNase Out (Life technologies)
DEPC water (Thermo Fisher Scientific) - Polysome buffer
50 mM Tris-HCl pH 7.5
5 mM MgCl2
25 mM KCl
0.2 M sucrose (Sigma-Aldrich)
100 µg/ml CHX (Sigma-Aldrich)
EDTA-free protease inhibitors (Roche)
10 U/ml RNase Out (Life technologies)
DEPC water (Thermo Fisher Scientific) - 15% sucrose solution
15% sucrose (m/v)
50 mM Tris-HCl pH 7.5
5 mM MgCl2
25 mM KCl
100 µg/ml CHX (Sigma-Aldrich)
EDTA-free protease inhibitors (Roche)
10 U/ml RNase Out (Life technologies)
DEPC water (Thermo Fisher Scientific) - 50% sucrose solution
50% sucrose (m/v)
50 mM Tris-HCl pH 7.5
5 mM MgCl2
25 mM KCl
100 µg/ml CHX (Sigma-Aldrich)
EDTA-free protease inhibitors (Roche)
10 U/ml RNase Out (Life technologies)
DEPC water (Thermo Fisher Scientific) - 3% PFA
Dilute stock solution (16%) in PBS 1x
Acknowledgments
JWY is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases. AD benefits from generous funding from Fondation pour la Recherche Médicale, Ligue contre le Cancer and Cancéropôle GSO. The authors declare no conflicts of interest or competing interests.
References
- Bastide, A., Peretti, D., Knight, J. R., Grosso, S., Spriggs, R. V., Pichon, X. Sbarrato, T., Roobol, A., Roobol, J., Vito, D., Bushell, M., von der Haar, T., Smales, C. M., Mallucci, G. R. and Willis, A. E. (2017). RTN3 is a novel cold-Induced protein and mediates neuroprotective effects of RBM3. Curr Biol CB 27: 638-650.
- David, A., Dolan, B. P., Hickman, H. D., Knowlton, J. J., Clavarino, G., Pierre, P., Bennink, J. R. and Yewdell, J. W. (2012). Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol 197(1): 45-57.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bastide, A., Yewdell, J. W. and David, A. (2018). Determining Ribosome Translational Status by Ribo-ELISA. Bio-protocol 8(1): e2670. DOI: 10.21769/BioProtoc.2670.
Category
Biochemistry > Protein > Immunodetection > ELISA
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









