- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Easy and Efficient Permeabilization of Cyanobacteria for in vivo Enzyme Assays Using B-PER
Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2667 Views: 7457
Reviewed by: Dennis NürnbergAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Lauren E.-A. Eyssen [...] Raymond J. Owens
Mar 20, 2024 6209 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2087 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2167 Views
Abstract
Cyanobacteria are photosynthetic bacteria that thrive in diverse ecosystems and play major roles in the global carbon cycle. The abilities of cyanobacteria to fix atmospheric CO2 and to allocate the fixed carbons to chemicals and biofuels have attracted growing attentions as sustainable microbial cell factories. A better understanding of activities of enzymes involved in the central carbon metabolism might lead to increased product yields. Currently, cell-free lysates are widely used for the determination of intracellular enzyme activities. However, due to thick cell walls in cyanobacteria, lysis of cyanobacterial cells is inefficient and often laborious. The present protocol describes an easy and efficient method to permeabilize cyanobacterial cells, without lysing them, and direct usage of the permeabilized cells for the determination of metabolic enzyme activities in vivo.
Keywords: B-PERBackground
We have previously reported an easy, efficient, and scalable permeabilization of cyanobacteria using the B-PERTM reagent (Thermo Fisher Science) (Rasmussen et al., 2016). The B-PERTM reagent contains a non-disclosed mild detergent dissolved in a Tris-HCl buffer and is typically used to lyse bacterial cells such as Escherichia coli. Serendipitously, we found that the B-PERTM reagent permeabilized cyanobacterial cells, instead of lysing them, likely because the thick cyanobacterial cell wall (Hoiczyk and Hansel, 2000) confers resistance to the detergent used in the reagent. Permeabilization was conducted in biotechnologically interesting cyanobacteria, Synechococcus sp. PCC 7002 (hereafter Synechococcus 7002) and Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803). Briefly, incubation of cyanobacterial cells in the B-PERTM reagent for 10 min resulted in permeabilization of the cells as confirmed by the SYTOX Green staining. No significant change in cell shape and no major loss of intracellular proteins were observed during the treatment. Determination of the activity of two enzymes, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and glucose-6-phosphate dehydrogenase (G6PDH), which play imperative roles in the central carbon metabolism, was performed. When used directly in the assays, the permeabilized cells exhibited the enzyme activities that were comparable to or even higher than those detected for cell-free lysates. Moreover, the permeabilized cells could be stored at -20 °C without losing the enzyme activities. The permeabilization process and subsequent activity assays were successfully adapted to a 96-well plate system, allowing mid-to-high throughput characterization. The protocol may be readily adapted to studies of other cyanobacterial species and other intracellular enzymes. The protocol presented in this article provides a standardized step-by-step procedure adapted to a 10-ml culture of exponentially-growing cyanobacteria, although it can also be scaled up to a larger culture volume (~1 L) or down to as little as 200-µl cultures grown in a microtiter plate (Rasmussen et al., 2016).
Materials and Reagents
- Eclipse® pipette tips (Labcon, catalog numbers: 4-1011-260-000 for 20 µl tips, 4-1018-260-000 for 200 µl tips, 4-1019-260-000 for 1,200 µl tips)
- 1-cm light path cuvettes, polystyrene/polymethyl methacrylate (VWR, catalog number: 634-0676 )
- NuncTM 96-well plate (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 249946 )
- 15-ml Falcon tubes (Greiner Bio One International, catalog number: 188271 )
- 1.5-ml Eppendorf safe-lock microtubes (Eppendorf, catalog number: 0030120086 )
- 1.5-ml Eppendorf microtubes (Eppendorf, catalog number: 0030125150 )
- 1.5-ml NalgeneTM screw cap microcentrifuge tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 342800-0020 )
- A culture of Synechococcus 7002 grown on a plate of A+ medium (Stevens et al., 1973) containing 1.5% (w/v) BD BactoTM dehydrated agar (BD, BactoTM, catalog number: 214050 )
- A culture of Synechocystis 6803 grown on a plate of BG11 medium (Stanier et al., 1971) containing 1.5% (w/v) BD BactoTM dehydrated agar (BD, BactoTM, catalog number: 214050 )
- A+ medium, liquid (Stevens et al., 1973)
- BG11 medium, liquid (Stanier et al., 1971)
- B-PERTM bacterial protein extraction reagent (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 78248 )
- A 20 mM Tris-HCl buffer, pH 7.5
- UltraPureTM Tris hydrochloride [tris(hydroxymethyl)aminomethane hydrochloride] (Tris-HCl) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15506017 )
- 96% (v/v) ethanol (Plum, catalog number: 201104 )
- 1 M sodium bicarbonate (NaHCO3) (Sigma-Aldrich, catalog number: S5761 )
- Micro BCATM protein assay kit (Thermo Fisher Scientific)
- cOmpleteTM protease inhibitor cocktail without ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, Roche Diagnostics, catalog number: 04693116001 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884 )
- Phosphocreatine (Sigma-Aldrich, catalog number: P7936 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014 )
- NADH (Sigma-Aldrich, Roche Diagnostics, catalog number: 10107735001 )
- Ribulose-1,5-bisphosphate (Sigma-Aldrich, catalog number: 83895 )
- Glyceraldehyde-3-phosphate dehydrogenase (Sigma-Aldrich, catalog number: G2267 )
- 3-Phosphoglyceric phosphokinase (Sigma-Aldrich, catalog number: P7634 )
- Creatine phosphokinase (Sigma-Aldrich, catalog number: C3755 )
- Tris(hydroxymethyl) aminoethane maleate (Sigma-Aldrich, catalog number: T3128 )
- NADP+ (Sigma-Aldrich, Roche Diagnostics, catalog number: 10128058001 )
- Glucose-6-phosphate (Sigma-Aldrich, Roche Diagnostics, catalog number: 10127647001 )
- cOmpleteTM protease inhibitor cocktail without ethylenediaminetetraacetic acid (EDTA) (see Recipes)
- Buffer A (see Recipes)
- A Rubisco assay buffer (see Recipes)
- G6PDH assay buffer (see Recipes)
Equipment
- Glass tubes (Inner diameter of 1.9 cm, max volume ~40 ml)
- P10 FinnpipetteTM F2 fixed volume single-channel pipettes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4642010 )
- P20 FinnpipetteTM F2 fixed volume single-channel pipettes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4642060 )
- P200 FinnpipetteTM F2 fixed volume single-channel pipettes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4642080 )
- P1000 FinnpipetteTM F2 fixed volume single-channel pipettes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4642090 )
- -20 °C freezer
- HiclaveTM autoclave (HMC Europe)
- Aquaria with temperature set at 30 °C for Synechocystis 6803 and 37 °C for Synechococcus 7002
- Philips Master TL-D, 18 W/840 cool white fluorescent tubes (Philips Lighting Holding, catalog number: 871150063171840 ). The number of fluorescence tubes is adjusted to achieve the photon flux of 50 µmol photons m-2 sec-1 for Synechocystis 6803 and 200 µmol photons m-2 sec-1 for Synechococcus 7002
- GMS150 gas mixer (Photon Systems Instruments, catalog number: GMS150 ) to provide 3% (v/v) CO2 balanced with air
- Holten horizontal laminar airflow sterile bench (Thermo Fisher Scientific, model: Holten Horizontal Laminar Airflow Clean Bench )
- QRT1 Quantitherm light meter/thermometer (Hansatech Instruments, model: QRT1 )
- Ultrospec 3100 pro UV/Visible spectrophotometer (GE Healthcare, Amersham Biosciences, model: Ultrospec 3100 pro )
- 2-L flask
- SorvallTM RC 6 Plus centrifuge (Thermo Fisher Scientific, model: SorvallTM RC 6 Plus , catalog number: 46910)
Note: This product has been discontinued. - PSU-20i multi-functional orbital shaking platform (Grant Instruments, model: PSU-20i )
- SpectraMax 190 microplate reader with SoftMax Pro software (Molecular Devices, model: SpectraMax 190 , catalog number: 190)
- Eppendorf ThermoMixer® C (Eppendorf, model: ThermoMixer® C , catalog number: 2231000269)
- Refrigerated tabletop centrifuge for 1.5 ml Eppendorf tubes (Thermo Fisher Scientific, Thermo ScientificTM, model: SorvallTM LegendTM Micro 17 , catalog number: 75002430)
Software
- Excel (Microsoft Office)
- SoftMax Pro software (see above for supplier)
Procedure
- Cultivation of cyanobacteria
- To make a pre-culture, transfer a small amount (approximately the size of a rice grain) of cyanobacterial cells from a plate culture into 10 ml of liquid medium. Grow the pre-culture in an aquarium (see Equipment, also see Figure 1) for 24 h with a constant supply of 3% (v/v) CO2 balanced with air. Use A+ medium and BG11 medium to grow Synechococcus 7002 and Synechocystis 6803, respectively.
- Transfer 1 ml of the culture into a plastic cuvette with a 1 cm light path length, and measure the optical density at 730 nm (OD730) using a spectrophotometer. If the value was higher than 0.8, make appropriate dilutions until the value falls between 0.1 and 0.8. Then calculate the OD730 value of the undiluted culture.
- Dilute the pre-culture with fresh media to achieve OD730 of 0.05 in the volume of 10 ml. Grow the culture until OD730 reaches approximately 1. At this stage, the cells are in the late exponential growth phase.
- Typically, three independent cultures are prepared as biological replicates for each condition. Examples of conditions are distinct genotypes (e.g., a wild-type strain, a mutant strain), nutrient regimens (e.g., deprivation of a micro/macro nutrient), and environmental setups (e.g., light intensity, temperature, pH).
- Durations needed for Synechococcus 7002 and Synechocystis 6803 cultures to reach OD730 of 1 from OD730 of 0.05 depend on conditions, but typically ~20 h and ~40 h, respectively, in the culture setups described in this protocol.
- The culture volume may be adjusted depending on needs. We tested up to 1 L in a 2-L flask and down to 200 μl in a 96-well plate.
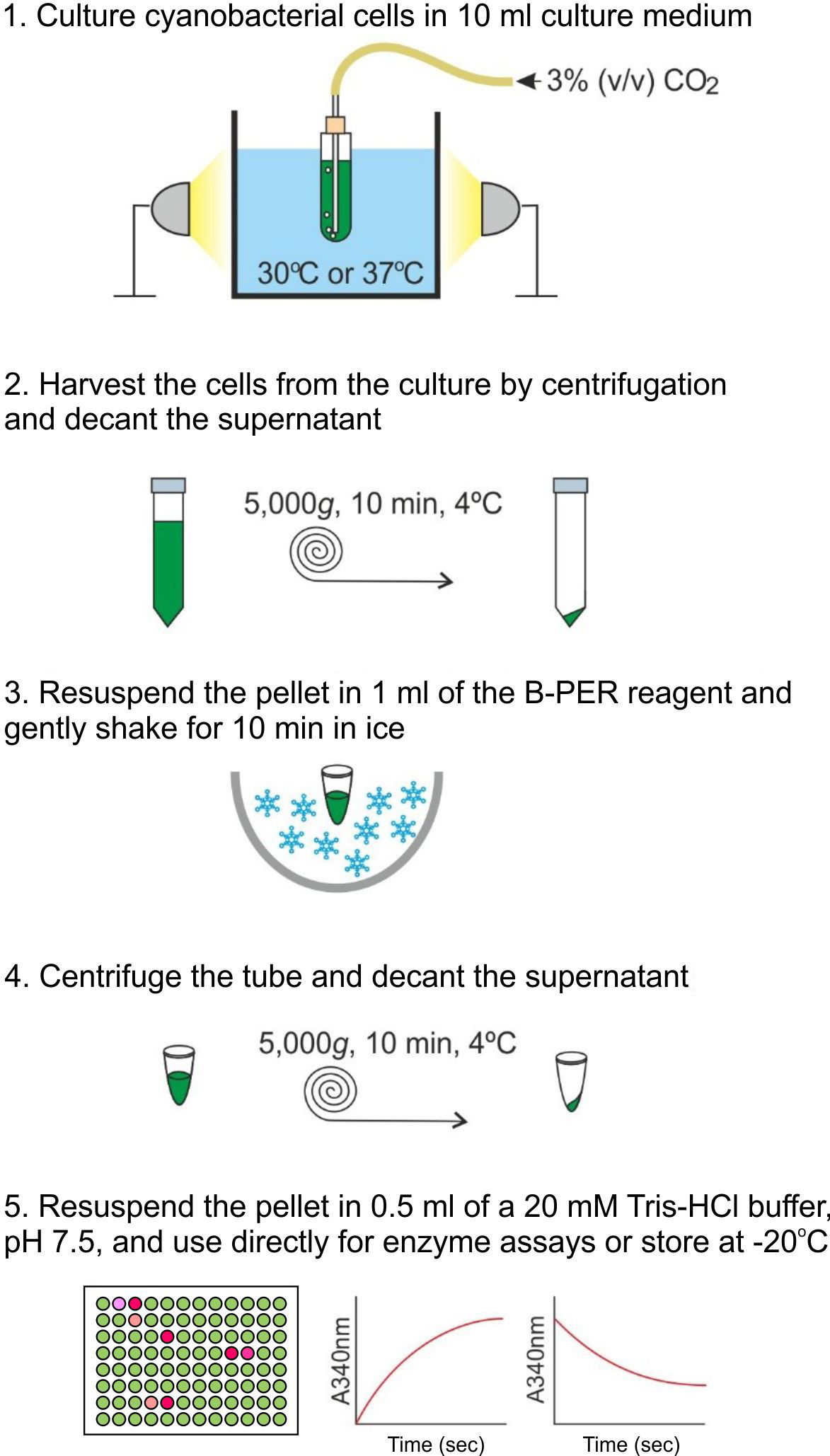
Figure 1. A schematic illustration of the procedure for cultivation, permeabilization, and enzyme activity assays of a 10-ml cyanobacterial culture - To make a pre-culture, transfer a small amount (approximately the size of a rice grain) of cyanobacterial cells from a plate culture into 10 ml of liquid medium. Grow the pre-culture in an aquarium (see Equipment, also see Figure 1) for 24 h with a constant supply of 3% (v/v) CO2 balanced with air. Use A+ medium and BG11 medium to grow Synechococcus 7002 and Synechocystis 6803, respectively.
- Permeabilization of cyanobacterial cells
Perform the following procedure for each of the biological replicates prepared in Procedure A.
Notes:- See Note a in Procedure A
- All steps are carried out on ice.
- See Figure 1.
- Prepare the following solutions and store on ice until use:
- The B-PERTM reagent containing protease inhibitors: add 40 µl of the 25x cOmplete protease inhibitor solution (see Recipes) to 1 ml B-PERTM reagent.
- A 20 mM Tris-HCl buffer, pH 7.5, containing protease inhibitors: add 20 µl of the 25x cOmplete protease inhibitor solution to 0.5 ml of a Tris-HCl buffer (20 mM, pH 7.5).
- The B-PERTM reagent containing protease inhibitors: add 40 µl of the 25x cOmplete protease inhibitor solution (see Recipes) to 1 ml B-PERTM reagent.
- Transfer the 10-ml culture, prepared in Step A3, to a 15 ml Falcon tube, and centrifuge at 5,000 x g for 10 min at 4 °C. Discard the supernatant.
Note: When a larger culture volume is used (e.g., 1 L), centrifuge bottles with a larger capacity (≥ 200 ml) would be suitable for harvesting cells. When a 96-well plate is used for cultivation, the plate can be directly centrifuged in a multi-well plate centrifuge. - Resuspend the pellet in 1 ml of the B-PERTM reagent containing protease inhibitors, prepared in Step B1a, by pipetting up and down until the cell suspension is homogenous.*
- Transfer the resuspension into a fresh 1.5-ml Eppendorf tube and shake gently for 10 min in ice using an orbital shaking platform at 100 rpm.
- Pellet the permeabilized cyanobacterial cells by centrifugation at 5,000 x g for 10 min at 4 °C. Discard the supernatant.*
- Resuspend the pellet containing permeabilized cyanobacterial cells in 500 µl of the Tris-HCl buffer containing protease inhibitors, prepared in Step B1b. Immediately use this resuspension for enzymatic assays or store at -20 °C for later use. See Figure 2 for a visual example of how treated cells look in comparison to untreated cells.

Figure 2. Treated and untreated cyanobacterial cells
- See Note a in Procedure A
- Determination of chlorophyll a concentrations
Perform the following procedure for each of the biological replicates prepared in Procedure A.- Briefly invert the permeabilized cyanobacterial cells prepared in Step B6 and transfer 100 μl in a 1.5-ml screw cap microcentrifuge tube.
Note: Permeabilized cells sediment quickly, so it is important to invert the tube once or twice before taking out the indicated volume. - Add 900 μl of 96% (v/v) ethanol and close the cap.
- Incubate the tube at 90 °C for 5 min with shaking at 700 rpm in a thermomixer.
- Cool down the tube on ice for 2 min.
Note: Cooling is important because the microtube becomes too hot to handle. - Spin down the cell debris at 10,000 x g for 10 min in a refrigerated tabletop centrifuge at 4 °C.
- Transfer the supernatant to a 1-cm light path cuvette.
- Measure the absorbance at 664 nm (A664) and 648 nm (A648) using a spectrophotometer.
Note: Measurements are performed typically within 5 min following the centrifugation step. - Calculate the chlorophyll a concentration as follows: Chlorophyll a (μg ml-1) = (13.36 x A664 - 5.19 x A648) x 10 (Lichtenthaler and Buschmann, 2001), wherein the multiplication factor of 10 is included to account for the dilution effect.
- Briefly invert the permeabilized cyanobacterial cells prepared in Step B6 and transfer 100 μl in a 1.5-ml screw cap microcentrifuge tube.
- Determination of enzymatic activities
Note: There are 3 biological replicates for each condition (see Note a in Procedure A); therefore, 9 wells are used for each condition (Steps D1a, D1c, D2a).- Rubisco activity assay
- Transfer 50 µl of the permeabilized cells prepared in B6 into a well in a 96-well plate. Perform this for each biological replicate.
Note: Permeabilized cells sediment quickly, so it is important to invert the tube once or twice before taking out the indicated volume. - Add 148 µl of buffer A (see Recipes) and 2 µl of 1 M NaHCO3 into each well containing the permeabilized cells and mix gently by pipetting up and down. Incubate on ice for 10 min to activate Rubisco by full carbamylation of the active site.
- Add 10 µl of the preincubated permeabilized cell preparation made in Step D1a to a well in a fresh 96-well plate. Prepare three technical replicates for each biological replicate.
- Add 240 µl of the Rubisco assay buffer (see Recipes) to each technical replicate and mix gently by pipetting up and down three times.
- Monitor the rate of NADH oxidation at a wavelength of 340 nm for 30 min using a microplate reader at 30 °C.
- Transfer 50 µl of the permeabilized cells prepared in B6 into a well in a 96-well plate. Perform this for each biological replicate.
- G6PDH activity assay
- Transfer 10 µl of the permeabilized cells prepared in B6 into a well in a 96-well plate. Prepare three technical replicates.
- Add 200 μl of a G6PDH assay buffer (see Recipes) to each technical replicate. Mix gently by pipetting up and down three times.
- Monitor the rate of formation of NADPH at a wavelength of 340 nm immediately after the addition of the assay buffer for 30 min using a plate reader at 30 °C.
- Transfer 10 µl of the permeabilized cells prepared in B6 into a well in a 96-well plate. Prepare three technical replicates.
- Rubisco activity assay
Data analysis
Calculate the Rubisco and G6PDH activities as follows:
- By using SoftMax Pro software, obtain a slope from each plot based on the linear initial rate period. Perform this for each technical replicate.
- Calculate the enzyme activities using the slope and the absorption coefficient for NADH/NADPH at 340 nm of 6.22 Abs mM-1 cm-1.
- Normalize the enzyme activities by the chlorophyll concentration.
Note: Protein concentrations and dry cell weights may be used, instead of the chlorophyll concentration, for normalizing enzyme activities. Typically, we use Micro BCATM protein assay kit (Thermo Fisher Scientific) for determining protein concentrations. For determination of dry cell weights, we use a previously described protocol (De Porcellinis et al., 2017). - Calculate the average value of the technical replicates. This represents the enzyme activity of a biological replicate.
- Calculate the average activity and standard deviation among three biological replicates. Student’s t-test or one-way ANOVA can be used to assess statistical significance of differences between conditions.
Notes
- When using B-PER permeabilization procedure, the enzymatic activities across independent experiments comparing the cells grown under the same conditions are reproducible.
- For high reproducibility, it is important to accurately weigh the correct amount of commercial enzymes used for enzymatic activity assays. The same principle applies when pipetting small volumes of substrates and other reagent to the enzymatic assays.
Recipes
- cOmpleteTM protease inhibitor cocktail without ethylenediaminetetraacetic acid (EDTA)
To make a 25x stock solution, one tablet is dissolved in 2 ml sterilized ddH2O and store at -20 °C - Buffer A
15 mM MgCl2
1 mM EDTA
50 mM Tris-HCl, pH 8.0 - A Rubisco assay buffer
50 mM Tris-HCl, pH 8.0
15 mM MgCl2
1 mM EDTA
5 mM phosphocreatine
10 mM NaCl
0.15 mM NADH
0.25 mM ribulose-1,5-bisphosphate
10 U glyceraldehyde-3-phosphate dehydrogenase
10 U 3-phosphoglyceric phosphokinase
2.5 U creatine phosphokinase (Stitt and Schulze, 1994) - G6PDH assay buffer
50 mM tris(hydroxymethyl) aminoethane maleate, pH 7.8
10 mM MgCl2
1 mM NADP+
10 mM glucose-6-phosphate (Schaeffer and Stanier, 1978)
Acknowledgments
The research leading to these results has received funding from the Innovation Fund Denmark (Grant ‘Plant Power’, No. 12-131834; Grant ‘Biomass for the 21st Century’, No. 001-2011-4), Nordic Energy Research (Grant ‘AquaFEED’, No. 24), European Union’s Seventh Framework Programme FP7-ENERGY-2010-1 under REA Grant agreement No. 256808 (Grant ‘DirectFuel’), the People Programme (Marie Curie Actions) FP7/2007-2013 under REA Grant agreement No. 317184 (Grant ‘PHOTO.COMM’). This protocol was adapted from our previous work (Rasmussen et al., 2016). There is no conflict of interest.
References
- De Porcellinis, A., Frigaard, N. U. and Sakuragi, Y. (2017). Determination of the glycogen content in cyanobacteria. J Vis Exp (125).
- Hoiczyk, E. and Hansel, A. (2000). Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol 182(5): 1191-1199.
- Lichtenthaler, H. K. and Buschmann, C. (2001). Chlorophylls and carotenoids: Measurement and characterization by UV-VIS. In: Current Protocols in Food Analytical Chemistry F4.3.1-F4. John Wiley and Sons.
- Rasmussen, R. E., Erstad, S. M., Ramos-Martinez, E. M., Fimognari, L., De Porcellinis, A. J. and Sakuragi, Y. (2016). An easy and efficient permeabilization protocol for in vivo enzyme activity assays in cyanobacteria. Microb Cell Fact 15(1): 186.
- Schaeffer, F. and Stanier, R. Y. (1978). Glucose-6-phosphate dehydrogenase of Anabaena sp. Kinetic and molecular properties. Arch Microbiol 116(1): 9-19.
- Stanier, R. Y., Kunisawa, R., Mandel, M. and Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35(2): 171-205.
- Stevens, S. E. Jr., Patterson, C. O. P. and Myers, J. (1973). The production of hydrogen peroxide by blue-green algae: a survey. J Phycol 9: 427-430.
- Stitt, M. and Schulze, D. (1994). Does Rubisco control the rate of photosynthesis and plant-growth? An exercise in molecular ecophysiology. Plant Cell Environ 17: 465-487.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Erstad, S. M. and Sakuragi, Y. (2018). Easy and Efficient Permeabilization of Cyanobacteria for in vivo Enzyme Assays Using B-PER. Bio-protocol 8(1): e2667. DOI: 10.21769/BioProtoc.2667.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









