- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Biomechanical Characterization of Onion Epidermal Cell Walls
Published: Vol 7, Iss 24, Dec 20, 2017 DOI: 10.21769/BioProtoc.2662 Views: 13192
Reviewed by: Manjula MummadisettiAlexandros AlexandratosAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analysis of Chromosome Condensation/Decondensation During Mitosis by EdU Incorporation in Nigella damascena L. Seedling Roots
Eugene V. Sheval
Feb 5, 2018 6900 Views

Visualization of Actin Organization and Quantification in Fixed Arabidopsis Pollen Grains and Tubes
Xiaolu Qu [...] Shanjin Huang
Jan 5, 2020 5342 Views

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2312 Views
Abstract
Here we describe two experimental protocols to measure the biomechanical properties of primary (growing) plant cell walls, with a focus on analyzing cell wall epidermal strips of onion scales. The first protocol measures cell wall creep (time-dependent irreversible extension) under constant force. Such creep is often mediated by the wall-loosening action of expansin or selective endoglucanases. The second protocol is based on two consecutive stretches of the wall and measures the wall’s elastic and plastic compliances, which depend on cell wall structure. These two assays provide complementary information that may be linked to cell wall structure and expansive growth of cells.
Keywords: Cell wallsBackground
The primary walls of growing plant cells are strong enough to resist the tensile forces generated by cell turgor pressure, yet can expand irreversibly during cell growth as a result of the selective loosening action of expansins or other catalysts necessary for irreversible cell wall enlargement (Cosgrove, 2016a and 2016b). Assessments of cell wall mechanical properties, such as elasticity and plasticity, are important for understanding cell wall structure and its modification during growth (cell enlargement) and after growth ceases. An important measure of wall mechanics is based on uniaxial tensile tests, as described below in our stress/strain assay, which measures elastic and plastic compliance (compliance is the reciprocal of modulus; modulus is a measure of stiffness). A second, complementary assay is based on the irreversible, time-dependent increase in length (creep) that occurs in primary cell walls when they are held at constant tension and continuously loosened by endogenous expansin or exogenous endoglucanase (Durachko and Cosgrove, 2009; Cosgrove, 2011; Cosgrove et al., 2017). In this protocol, we describe procedures for preparing epidermal cell wall strips from onion scales and testing them in these two assays. Onion epidermal cell walls provide a useful model to explore the connection between cell wall structure and biomechanics (Wilson et al., 2000; Suslov et al., 2009; Kim et al., 2015; Zhang et al., 2017; Zheng et al., 2017).
Materials and Reagents
- Single edge razor blades (e.g., VWR, catalog number: 55411-050 )
- Double edge razor blades (e.g., Safety Razor Company, catalog number: 74-0002 )
- Disposable Petri dishes (e.g., Corning, Falcon®, catalog number: 351007 )
- Double sided ½ inch wide ‘Scotch’ tape
- Masking tape or similar (e.g., VWR, catalog number: 89097-920 )
- 25 x 75 mm, 1.0 mm thick microscope slides (e.g., VWR, catalog number: 48300-025 )
- 22 x 22-1.5 microscope cover glass (e.g., Fisher Scientific, Fisherbrand, catalog number: 12-541-B )
- White onion (Allium cepa L.) bulbs; we use the Cometa cultivar
- Distilled-deionized water (ddH2O)
- Sodium acetate trihydrate (e.g., VWR, catalog number: BDH9278-500G )
- HEPES (e.g., VWR, catalog number: 97061-826 )
- Hydrochloric acid (e.g., Merck, catalog number: HX0603-75 )
- Acetic acid (e.g., Merck, catalog number: AX0073-6 )
- HEPES buffer (20 mM, pH 6.8) (see Recipes)
- Sodium acetate buffer (20 mM, pH 4.5) (see Recipes)
Equipment
- Microwave oven, hot plate, or Bunsen burner
- pH meter (e.g., Corning, model: Model 430 )
- Analytical balance (e.g., Mettler Toledo, model: XPE105 )
- Fine forceps (e.g., Dumont No. 5)
- Flat tip forceps (e.g., Rose Scientific, catalog number: FF-001 )
- Custom 3 mm slicing jig (see Notes section)
- Microcomputer
- Data acquisition hardware (e.g., Data Translation, model: DT9801 )
- Constant-force extensometer (Figure 1) (Durachko and Cosgrove, 2009; Cosgrove, 2011; Cosgrove et al., 2017)
- Stress/strain analyzer (Figure 2) (Cosgrove, 2011; Cosgrove et al., 2017)

Figure 1. The constant-force extensometer. A vertically adjustable, open clamping chamber fixes the bottom of the wall sample and is capable of holding liquid with a capacity of roughly 150 microliters. A second clamp is applied to the upper portion of the wall sample and is connected to a balanced lever. Displacement of the lever and connecting rod is monitored using a linear variable displacement transducer (LVDT), e.g., Schaevitz 050HR. Weights are applied to the lever to modulate the tension on the sample. Custom software displays and records displacement and calculated extension rates. We use a bank of eight of these units connected in parallel to a microcomputer via a data acquisition module (Data Translation DT9801).
Figure 2. The device used for stress/strain assays. A. The vertical stage is controlled by a PC serial port stepper driver (e.g., AutomationDirect STP-DRV-4850), stepper motor (e.g., Lin Engineering 4218L-01D-02), geared belt drive, and lead screw assembly. Stage position is monitored with an LVDT. Tension is monitored with an s-type load cell (e.g., Futek LSB200 100 g). Custom software controls the strain rate and tension. Two clamps hold the sample being measured. The upper clamp is fixed to the movable stage. The lower clamp is fixed to the load cell. B. Close up of peel affixed between clamps (peel stained blue to aid visualization for this image). C. Alternate view of device noting specific pieces.
Software
- Custom software written using Microsoft Visual Basic is used for acquiring data and some data analysis. Microsoft Visual Basic may be freely downloaded at http://www.microsoft.com. Alternatively, many commercial software packages are available both free and for purchase to allow data acquisition and analysis
Procedure
- Onion peel preparation
Here we describe the protocol for preparing a cell wall strip from the onion. This is obtained from the abaxial epidermis of onion scales (from the outer or convex surface of the scale). The abaxial epidermis adheres tightly to the underlying cells and so when one makes a peel the outer (periclinal) epidermal wall separates from the rest of the cell, resulting in a strip made up of the outer wall only. This contrasts with the peeling behavior of the adaxial epidermis (from the inner or concave surface of the onion scale), which is only weakly attached to the underlying cells and consequently separates as a whole-cell layer with cells living and intact, e.g., Hepworth and Bruce, 2004; Vanstreels et al., 2005; Suslov et al., 2009; Beauzamy et al., 2015.- Purchase fresh white onions roughly the size of a baseball (between 6 and 10 cm in diameter).
- Remove and discard the outermost dry, brown layers (Figure 3A).
- The remainder will be readily separable into layers called scales. We refer to these scales from outermost (oldest) to innermost by number (outer to inner, 1, 2..., n) (Figure 3B).
- Use a single edge razor blade to slice the onion vertically to obtain tapered scale sections of roughly 3 to 4 cm at their widest dimension (Figure 3B).
- Remove scales 1 through 4, leaving scale 5 on the onion from which we obtain our peels. It is also possible to make peels from the other scales.
- Use the 3 mm slicing jig to make shallow (~0.5 mm deep, 3 mm wide) incisions on the outer (abaxial) surface of scale #5 along the onion vertical axis. These cuts should be roughly 3 cm long and are centered over the middle of the onion vertical axis (Figure 3C).
- Use a single edge razor blade to make three similar incisions perpendicular to existing ones. These three cuts should be roughly 1 cm apart and centered about the onion vertical axis. This allows for four peels per set of incisions (Figure 3D, lines indicate incisions, one peel per rectangle).

Figure 3. Preparing an onion for obtaining peel strips. A. Remove dry layers; B. Excise to scale #5; C. Make vertical incisions; D. Make horizontal incisions. - Using the flat tip forceps, gently work under one end of the four incised rectangles around 0.5 mm deep into the subdermal tissue. Pinch this tissue and use it to gently peel off the epidermal layer approximately halfway along the long axis of one of your four incised rectangles. Then, working from the other end of the rectangle, peel the remainder to get an intact rectangle of epidermal tissue with chunks of subdermal tissue adhering to both ends. Float peels on HEPES buffer in a Petri dish (Figure 4).

Figure 4. Peeling strips from the onion (A) and floating them on HEPES buffer (20 mM, pH 6.8) (B) - Strips ~1 cm long and 3 mm wide are now ready for use in biomechanical assays. For the assays described in this paper, we clamp both ends into one of two devices, with an initial length of 3 or 5 mm between clamps.
- Purchase fresh white onions roughly the size of a baseball (between 6 and 10 cm in diameter).
- Constant-force extensometer (creep) assay
This is the procedure for measuring cell wall creep, e.g., mediated by catalysts for cell wall loosening (Cosgrove, 2016a).- Ensure all units of extensometer are clean and ready for a new experiment.
- We typically use 15 to 17 g of tension (0.15-0.17 N) for 3 mm wide abaxial onion epidermal peels from scale #5. Wall strips from younger and older scales have different properties, so you may need to adjust the extensometer tensioning system by trial and error (sufficient tension to obtain good creep rates, not so much that wall breakage becomes a problem).
- Prepare an appropriate number of peels and proceed with loading as many units as you desire for your particular experiment. We find 8 to 16 replicates to be typically adequate to account for sample variability.
- Heat inactivation
Note: Steps B4a-B4g are only used when one wants to inactivate endogenous wall enzymes and they occur before loading the peels in the extensometer.- Place the peels between microscope slides and secure with a rubber band (Figure 5).

Figure 5. Three peels between slides secured with a rubber band - Put peels in slides into a heat-safe vessel with 100 ml of distilled water so that they can lay flat and be completely submerged.
- Place into a microwave oven on its highest setting and start the oven.
- Watch carefully and once a rolling boil is established allow samples to heat for 12 sec.
- Remove immediately and quench by placing the slides with peels into another vessel containing room temperature water.
- A Bunsen burner or hot plate may be used but alter the procedure as follows: allow the water to come to a boil and then submerge the peels in slides into the already boiling water. After 12 sec quench as for the microwave protocol.
- Once the microscope slides are cool, place the peels into buffer as dictated by experimental requirements.
- Place the peels between microscope slides and secure with a rubber band (Figure 5).
- Remove one peel from the Petri dish and place it onto a cover glass, which serves as a carrier for the peel (Figure 6A).
- Position the peel at the top center of the cover glass (Figure 6B).
- Open the upper clamp with one hand.
- Insert one end of the peel into the upper clamp of the extensometer and clamp it (Figure 6C).
- Slide the cover glass down away from the upper end of the peel so as to allow it to hang within the lower clamp. Position the sample so as to clamp a 5 mm length of tissue between the clamps (Figure 6D).
- Clamp the lower clamp onto the peel.
- Add buffer to the freshly hung sample as dictated by experimental requirements (Figure 6E).

Figure 6. Procedures of a uniaxial wall extension (creep) assay. A. Transfer peel from solution to cover glass; B. Position peel at top center of cover glass; C. Insert peel into upper clamp and remove cover glass; D. Insert peel into lower clamp; E. Add buffer to peel. - Adjust the lower clamp assembly so as to ‘zero’ the displacement transducer into the lower portion of its range.
- Proceed with remaining empty units.
- Begin acquiring length data.
- After recording stable baseline rates, solutions covering the samples are exchanged as dictated by experimental needs, e.g., different pH or addition of enzymes such as expansin (Cosgrove, 2015 and 2016a) or endoglucanases (Park and Cosgrove, 2012) (Figure 7).

Figure 7. Uniaxial cell creep extension of native onion epidermal cell wall induced by acidic buffer. Native wall strip (that is, not inactivated by heat) clamped at 10 g tension exhibits acid-induced creep when the initial neutral buffer (20 mM HEPES, pH 6.8) is exchanged for 20 mM sodium acetate, pH 4.5, at the point indicated by the arrow.
- Ensure all units of extensometer are clean and ready for a new experiment.
- Stress/strain assay
Here we describe a protocol for quantifying the cell wall mechanical properties based on stress/strain curves (Cosgrove et al., 2017). The wall is stretched until a defined force is attained, then returned to original length and stretched a second time to the same force. The second stretch is reversible and thus provides a way to measure wall elasticity. The first stretch is partly irreversible (plastic) and so may be combined with the second stretch to calculate plasticity. - Adjust the extensometer software for a strain rate of 3 mm per minute, which is continued until a tension of 98 mN is reached. This tension may need to be smaller or larger, depending on the treatment and history of the cell wall strip. It should be set low enough to avoid tearing of the wall strip or slipping in the clamp, but large enough to produce plastic extension.
- We configure our device to use a starting distance of either 3 or 5 mm between clamps. This length of sample is convenient and provides sufficient material from which we can obtain a magnitude of measurable biomechanical response we require for these procedures.
- Prepare an appropriate number of peels as needed for your particular experiment. We typically find 12 to 15 replicates to be adequate to account for sample variability.
- Steps C4a-C4g are only used when one wants to inactivate endogenous wall enzymes.
- Place the peels between microscope slides and secure with a rubber band.
- Put peels in slides into a heat-safe vessel with 100 ml of distilled water so that they can lay flat and be completely submerged.
- Place into a microwave oven on its highest setting and start the oven.
- Watch carefully and once a rolling boil is established allow samples to be heated for 12 sec.
- Remove immediately and quench by placing the slides with peels into another vessel containing room temperature water.
- A Bunsen burner or hot plate may be used but alter the procedure as follows: allow the water to come to a boil and then submerge the peels in slides into the already boiling water. After 12 sec quench as for the microwave protocol.
- Once the microscope slides are cool, place the peels back into buffer as dictated by experimental requirements.
- Place the peels between microscope slides and secure with a rubber band.
- Remove one peel from the Petri dish and place it onto a cover glass, which serves as a carrier for the peel (Figure 6A).
- Position the peel at the top center of the cover glass (Figure 6B).
- Open the upper clamp with one hand.
- Refer to Figure 6 panels C and D for Steps 14 through 16 below. The device clamps are similar.
- Insert one end of the peel into the upper clamp of the device and clamp it.
- Slide the cover glass down away from the upper end of the peel so as to allow it to hang within the lower clamp.
- Clamp the lower clamp onto the peel.
- Adjust the device to take any slack out of the sample.
- Stretch the sample twice consecutively until a stress of 98 mN is reached each time (see Figure 8 and Videos 1-3 for examples of what the stretching looks like).
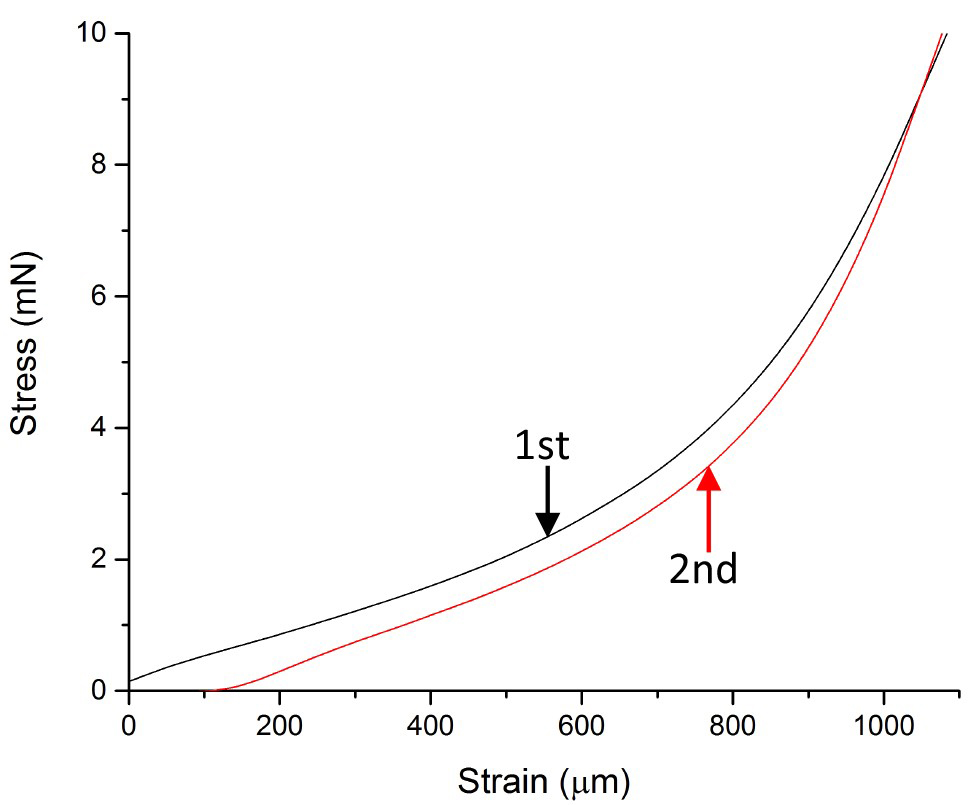
Figure 8. Representative stress-strain (force-extension) measurement. An onion epidermal cell wall strip was stretched in two cycles to a 98 mN load twice at a strain rate of 3 mm min-1.
Here are three movies illustrating the two sequential extension cycles for a stress/strain assay of onion epidermal wall strips.Video 1. An example of a moderately stiff wallVideo 2. An example of a less-stiff wallVideo 3. An example with slippage at the top clamp (this is an error condition)
Data analysis
- Extensometer data is gathered as follows: For each displacement transducer a position sample is captured every 30 sec. Those thirty samples are then averaged and saved to a data file as a single average position data point. Each single extension rate data point is calculated from 3 consecutive position points and saved to the same data file.
- For stress/strain analysis the slope of each stress/strain line is calculated by fitting the final 10% of the data gathered up until the target stress is reached using a linear least squares fit. The slope of the line (% extension versus force) is known as a compliance (the reciprocal of stiffness or modulus).
- The slope of the second stretch represents the elastic compliance (% extension/change in tensile force). The slope of the first stretch represents the total (elastic + plastic) compliance. The plastic compliance may be obtained by subtracting the elastic compliance from the total compliance.
Notes
- Incubation times for peels should always be minimized to attenuate any changes which might be caused by endogenous biological activities, unless that is the aim of the experiment. We also keep our samples on ice prior to use whenever it seems appropriate.
- Making the custom 3 mm slicing jig (Figure 9): This jig makes 3 mm wide peels. You will need three double edge razor blades, double stick ‘Scotch’ tape, masking tape (or similar) and 8 standard microscope slides (25 x 75 mm, 1.0 mm thick). On a clean, flat surface lay out the slides and razor blades. Put two pieces of double stick tape side-by-side on the upper surface of each slide to almost completely cover the slide surface. Place one blade near the end of a slide keeping a cutting edge parallel to the slide and protruding 5 mm from the edge of the slide. Place three slides on top of the first blade. Follow with another blade, 3 more slides, the final blade, and then the final slide. Be careful to assemble the parts in an orthogonal manner. You may wish to wrap the ends of the jig with additional tape.

Figure 9. Custom 3 mm slicing jig - When performing stress/strain analyses proceed at a pace so as not to allow tissue samples to dehydrate. Small amounts of buffer may be applied using a cotton-tipped swab or pipet if needed after clamping tissue and prior to collecting data.
Recipes
- HEPES buffer (20 mM, pH 6.8)
Dissolve 4.77 g of (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) in 0.95 L of ddH2O, adjust to pH 6.8 with 1 N HCl, and add ddH2O to make a final volume of 1 L - Sodium acetate buffer (20 mM, pH 4.5)
Dissolve 1.64 g of sodium acetate trihydrate in 0.95 L ddH2O and adjust to pH 4.5 with 1 M acetic acid, and add ddH2O to make a final volume of 1 L
Acknowledgments
We thank Ed Wagner, Dr. Sarah Kiemle and Xuan Wang for technical assistance. This work was supported by the Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences (grant No. DE-SC0001090). This protocol is adapted from Zhang et al. (2017). The authors declare no conflicts of interest or competing interests.
References
- Beauzamy, L., Derr, J. and Boudaoud, A. (2015). Quantifying hydrostatic pressure in plant cells by using indentation with an atomic force microscope. Biophys J 108(10): 2448-2456.
- Cosgrove, D. J. (2011). Measuring in vitro extensibility of growing plant cell walls. Methods Mol Biol 715: 291-303.
- Cosgrove, D. J. (2015). Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25: 162-172.
- Cosgrove, D. J. (2016a). Catalysts of plant cell wall loosening. F1000Res 5.
- Cosgrove, D. J. (2016b). Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot 67(2): 463-476.
- Cosgrove, D. J., Hepler, N. K., Wagner, E. R. and Durachko, D. M. (2017). Measuring the biomechanical loosening action of bacterial expansins on paper and plant cell walls. Methods Mol Biol 1588: 157-165.
- Durachko, D. M. and Cosgrove, D. J. (2009). Measuring plant cell wall extension (creep) induced by acidic pH and by alpha-expansin. J Vis Exp(25): 1263.
- Hepworth, D. G. and Bruce, D. M. (2004). Relationships between primary plant cell wall architecture and mechanical properties for onion bulb scale epidermal cells. J Texture Stud 35: 586-602.
- Kim, K., Yi, H., Zamil, M. S., Haque, M. A. and Puri, V. M. (2015). Multiscale stress-strain characterization of onion outer epidermal tissue in wet and dry states. Am J Bot 102(1): 12-20.
- Park, Y. B. and Cosgrove, D. J. (2012). A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol 158(4): 1933-1943.
- Suslov, D., Verbelen, J. P. and Vissenberg, K. (2009). Onion epidermis as a new model to study the control of growth anisotropy in higher plants. J Exp Bot 60(14): 4175-4187.
- Vanstreels, E., Alamar, A. C., Verlinden, B. E., Enninghorst, A., Loodts, J. K. A., Tijskens, E., Ramon, H. and Nicolai, B. M. (2005). Micromechanical behaviour of onion epidermal tissue. Postharvest Biol Tec 37: 163-173.
- Wilson, R. H., Smith, A. C., Kacurakova, M., Saunders, P. K., Wellner, N. and Waldron, K. W. (2000). The mechanical properties and molecular dynamics of plant cell wall polysaccharides studied by Fourier-transform infrared spectroscopy. Plant Physiol 124(1): 397-405.
- Zhang, T., Vavylonis, D., Durachko, D. M. and Cosgrove, D. J. (2017). Nanoscale movements of cellulose microfibrils in primary cell walls. Nat Plants 3: 17056.
- Zheng, Y., Cosgrove, D. J. and Ning, G. (2017). High-resolution field emission scanning electron microscopy (FESEM) imaging of cellulose microfibril organization in plant primary cell walls. Microsc Microanal 23(5): 1048-1054.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Durachko, D. M., Park, Y. B., Zhang, T. and Cosgrove, D. J. (2017). Biomechanical Characterization of Onion Epidermal Cell Walls. Bio-protocol 7(24): e2662. DOI: 10.21769/BioProtoc.2662.
Category
Plant Science > Plant cell biology > Cell structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











