- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Oral Microbiome Characterization in Murine Models
Published: Vol 7, Iss 24, Dec 20, 2017 DOI: 10.21769/BioProtoc.2655 Views: 10963
Reviewed by: Ivan ZanoniYang FuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Visualizing Hypoxia in a Murine Model of Candida albicans Infection Using in vivo Biofluorencence
José Pedro Lopes and Constantin F. Urban
Aug 5, 2019 5103 Views

RNA Extraction from Ears and Draining Lymph Nodes of Mice Infected with Leishmania amazonensis
Emilie Giraud and Evie Melanitou
Jun 5, 2020 5513 Views

TetR Regulated in vivo Repression Technology to Identify Conditional Gene Silencing in Genetically Engineerable Bacteria Using Vibrio cholerae Murine Infections as Model System
Franz G. Zingl [...] Stefan Schild
Oct 5, 2020 3742 Views
Abstract
The oral microbiome has been implicated as a trigger for immune responsiveness in the oral cavity, particularly in the setting of the inflammatory disease periodontitis. The protocol presented here is aimed at characterizing the oral microbiome in murine models at steady state and during perturbations of immunity or physiology. Herein, we describe murine oral microbiome sampling procedures, processing of low biomass samples and subsequent microbiome characterization based on 16S rRNA gene sequencing.
Keywords: Oral microbiomeBackground
The microbiome plays critical roles in modulating tissue-specific immune responses particularly at barrier sites (Belkaid and Harrison, 2017). In these barrier environments, such as the gastrointestinal tract and skin, select commensals are shown to be able to drive the development of specific immune cell populations (Ivanov et al., 2009; Naik et al., 2012). Our work has recently started to explore the influence of the oral microbiome in tailoring tissue immunity, particularly at the gingiva, a vulnerable oral barrier site (Abusleme and Moutsopoulos, 2016; Dutzan et al., 2017).
In humans, it is well recognized that the oral cavity harbors a diverse and rich microbiome (Human Microbiome Project, 2012). Alterations in oral microbial communities have been associated with the common oral disease, periodontitis, an inflammatory condition that affects the gingival tissues and results in tissue damage (Griffen et al., 2012; Abusleme et al., 2013; Moutsopoulos et al., 2015). To date, animal models have been instrumental in addressing the role of the microbiome in various physiological and pathological conditions (Turnbaugh et al., 2006; Kostic et al., 2013). However, studies of host-microbiome interactions have been increasingly challenging in the oral murine setting. To facilitate oral microbiome studies in murine models, we have developed protocols for sampling of oral microbial communities, processing of low biomass murine oral microbiome samples, 16S rRNA gene sequencing and analysis of relevant data.
Materials and Reagents
- Oral and gingival sample collection
- Ultra-Fine polystyrene swab (Puritan Medical Products, catalog number: 25-800 1PD 50 )
- Safe-lock Biopur 1.5 ml Individually wrapped tubes (Eppendorf, catalog number: 022600028 )
- KIMWIPES delicate task wipers (KCWW, Kimberly-Clark Professional, catalog number: 34120 )
- Sterile scalpel handle #3 (Fine Science Tools, catalog number: 10003-12 )
- Scalpel blades #10 (Fine Science Tools, catalog number: 10010-00 )
- 70% ethanol
- TE buffer (Epicentre, catalog number: MTE0970 )
- RNase AWAY decontamination reagent (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10328011 )
- Ketamine 100 mg/ml (Zetamine CIII, VetOne)
- Xylazine 20 mg/ml (AnaSed, Lloyd laboratories)
- Ultra-Fine polystyrene swab (Puritan Medical Products, catalog number: 25-800 1PD 50 )
- DNA isolation
- Safe-lock Biopur 1.5 ml Individually wrapped tubes (Eppendorf, catalog number: 022600028 )
- DNA IQ spin baskets (Promega, catalog number: V1225 )
- DNeasy Blood and Tissue Kit (QIAGEN, catalog number: 69504 )
- Ready-Lyse lysozyme solution (Epicentre, catalog number: R1810M )
- Ethanol for molecular biology (Sigma-Aldrich, catalog number: E7023 )
- RNase AWAY decontamination reagent (Thermo Fisher Scientific, catalog number: 10328011 )
- Safe-lock Biopur 1.5 ml Individually wrapped tubes (Eppendorf, catalog number: 022600028 )
- 16S rRNA gene library amplification and visualization
- DNA low-bind tube 1.5 ml (Eppendorf, catalog number: 0030108051 )
- DNA low-bind tube 2.0 ml (Eppendorf, catalog number: 0030108078 )
- PCR tubes 0.2 ml without caps (Bio-Rad Laboratories, catalog number: TLS0801 )
- PCR tube 8-Cap strips (Bio-Rad Laboratories, catalog number: TCS0803 )
- Platinum Taq DNA polymerase high fidelity (Thermo Fisher Scientific, catalog number: 11304011 )
- dNTP mix (10 mM each) (Thermo Fisher Scientific, catalog number: R0191 )
- PCR water (QIAGEN, catalog number: 17000-10 )
- Forward primer 8F 5’-AGAGTTTGATCMTGGCTCAG-3’ (Custom order–IDT)*
- Reverse primer 361R 5’-CYIACTGCTGCCTCCCGTAG-3’ (Custom order–IDT)*
*Note: These universal primers were first described in the article by Sundquist et al. (2007). Additionally, both forward and reverse primers need to include 5’ and 3’ linker sequences, heterogeneity spacers and the desired index identifier sequences that will allow dual indexing for later sample identification. These modifications are described in detail in the study of Fadrosh et al. (2014). - UltraPure agarose (Thermo Fisher Scientific, catalog number: 16500500 )
- 50x TAE buffer (Bio-Rad Laboratories, catalog number: 1610743 )
- GelPilot DNA Loading dye, 5x (QIAGEN, catalog number: 239901 )
- UltraPure ethidium bromide (Thermo Fisher Scientific, catalog number: 15585011 )
- 100 bp DNA ladder (New England BioLabs, catalog number: N3231S )
- RNase AWAY decontamination reagent (Thermo Fisher Scientific, catalog number: 10328011 )
- DNA low-bind tube 1.5 ml (Eppendorf, catalog number: 0030108051 )
- PCR Product Clean-up, quantification and sequencing
- DNA low-bind tube 1.5 ml (Eppendorf, catalog number: 0030108051 )
- Qubit assay tubes (Thermo Fisher Scientific, catalog number: Q32856 )
- Agencourt AMPure XP (Beckman Coulter, catalog number: A63880 )
- Ethanol for molecular biology (Sigma-Aldrich, catalog number: E7023 )
- Buffer EB (QIAGEN, catalog number: 19086 )
- Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, catalog number: Q32851 )
- Experion DNA 1K Kit (Bio-Rad Laboratories, catalog number: 7007107 )
- PhiX Control V3 (Illumina, catalog number: FC-110-3001 )
- MiSeq Reagent Kit v3 (Illumina, catalog number: MS-102-3003 )
- RNase AWAY decontamination reagent (Thermo Fisher Scientific, catalog number: 10328011 )
- DNA low-bind tube 1.5 ml (Eppendorf, catalog number: 0030108051 )
Equipment
- Sterile scissors (VWR, catalog number: 82027-582 )
- Sterile Fine scissors (Fine Science Tools, catalog number: 14058-11 )
- Sterile medium tipped forceps (Fisher Scientific, catalog number: 12-000-157 )
- Pipettes
- UV crosslinker (UV Stratalinker 1800) (Stratagene, model: Model 1800 )
- Microcentrifuge (Eppendorf, model: 5424 R )
- Heated shaking block for Eppendorf tubes (Eppendorf, model: Thermomixer 5436 )
- Vortex mixer (Scientific Industries, model: Vortex-Genie 2 )
- Spectrophotometer (Thermo Fisher Scientific, model: NanoDropTM 1000 , catalog number: ND-1000)
- Thermal Cycler (Bio-Rad Laboratories, model: DNA Engine )
- Electrophoresis Chamber (Bio-Rad Laboratories, model: Wide Mini-Sub Cell GT Cell )
- Gel Imager (Syngene, model: InGenius3 )
- DynaMag-2 Magnet (Thermo Fisher Scientific, model: DynaMagTM-2 Magnet )
- Automated electrophoresis station (Bio-Rad Laboratories, model: ExperionTM Automated Electrophoresis System )
- Experion priming station (Bio-Rad Laboratories, catalog number: 7007030 )
- Experion vortex station II (Bio-Rad Laboratories, catalog number: 7007043 )
- Qubit 3.0 Fluorometer (Thermo Fisher Scientific, model: Qubit 3.0 )
Software
- Software Mothur (Schloss et al., 2009)
- Software R (R Core Team, 2017)
- Software RStudio (RStudio Team, 2015)
Procedure
- Oral and gingival sample collection
- Oral mucosal microbiome sampling
Note: This procedure allows for comprehensive evaluation of microbiome from all oral mucosal areas.- Aseptically add 150 µl of TE buffer to 1.5 ml Safe-lock Biopur tubes. Include one per mouse to be sampled and one negative control.
- Clean work surface with 70% ethanol and RNase AWAY reagent. UV-treat tubes, racks, scissors for 30 min.
- Anesthetize or euthanize a mouse according to ARAC guidelines.
Note: For anesthesia, we prepare a liquid solution consisting of Ketamine (100 mg/kg) and Xylazine (10 mg/kg), which is administered via intraperitoneal injection. We recommend that each mouse is weighed prior to injection, for dosing calculation. - Wipe gloved hands with a tissue wipe previously moistened with RNase AWAY reagent.
- Open swab envelope and have it ready (one hand), grab the mouse pulling the skin of the neck area so the mouth opens without touching it (other hand) (Figure 1A).
- Swab the oral cavity for 30 sec, starting on the tongue, then buccal areas, gingiva, palate and finish with the gingiva on the lower incisors (Figure 1B).
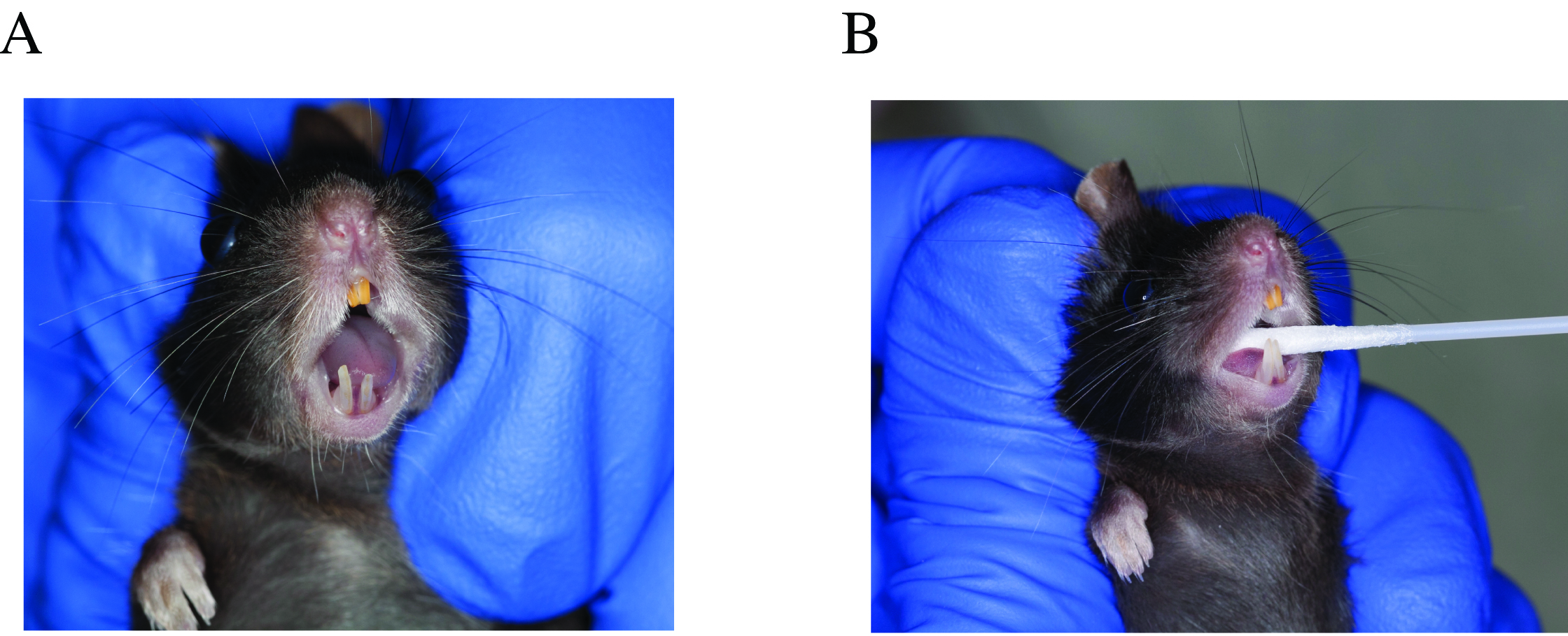
Figure 1. Oral swab sample collection. A. Hand positioning when handling the mouse for sampling, opening its mouth without touching it. B. Swab placement in the oral cavity, sampling tongue and buccal surfaces. - Place the swab in TE buffer and cut the swab handle with scissors, to allow for the tube to close, but not leaving it too short. Place the tube immediately on dry ice. Collect a negative control swab, by taking a swab and exposing it to the air and then placing it into a tube with TE buffer.
- After collecting all samples, store them at -80 °C until processing.
- Aseptically add 150 µl of TE buffer to 1.5 ml Safe-lock Biopur tubes. Include one per mouse to be sampled and one negative control.
- Gingival tissue sample collection
Note: This procedure allows for evaluation of the oral microbiome at the gingiva and may be most useful in studies of periodontitis.- Steps A2a-A2b as A1a-A1b.
- Euthanize a mouse according to ARAC guidelines.
Note: For euthanasia, we expose the mice to CO2 in an appropriate chamber, following the guidelines established by the Institutional Animal Care and Use Committee, NIDCR/NIH. - Wipe gloved hands with a tissue wipe previously moistened with RNase AWAY reagent.
- Perfuse the mouse and dissect tissues to expose the oral cavity (Figure 2A). These procedures are explained in detail in Dutzan et al. (2016).
- Using a #10 blade, dissect the palatal gingiva around the maxillary molars (Figure 2B). Remove gingival tissue with forceps and place it in a tube with TE buffer, which is directly transferred on dry ice. As a negative control, dip an unused pair of forceps in another tube with TE buffer.
- After collecting all samples, store them at -80 °C until processing.
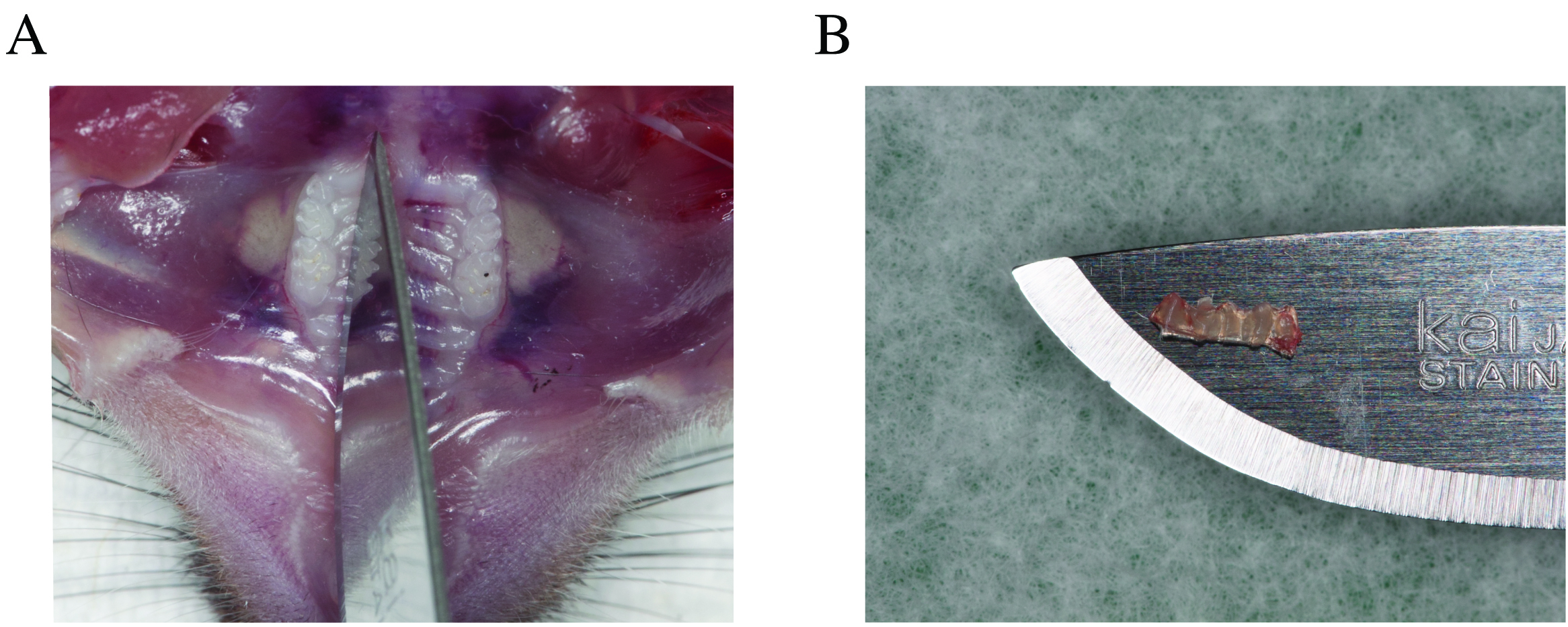
Figure 2. Gingival tissue collection. A. Dissection of palatal gingival tissue around molar teeth from mouse maxilla. B. Enlarged view of the excised gingival tissue.
- Steps A2a-A2b as A1a-A1b.
- Oral mucosal microbiome sampling
- DNA isolation
- Clean work surface with 70% ethanol and then RNase AWAY reagent. UV-treat a plastic tube rack, pipettes, sterile forceps and spin baskets (consider one pair of forceps and one basket per sample).
- Thaw previously collected samples by placing the tubes at 37 °C for 10 min on a heated shaking block at moderate speed.
- Briefly, spin sample tubes and add 1 µl of ReadyLyse per tube. Incubate for 60 min on a heated shaking block (37 °C) at moderate speed.
- Briefly spin tubes before opening.
- Add 25 µl of Proteinase K and 200 µl of buffer AL (both from the DNeasy Blood and Tissue Kit).
- Mix by vortexing briefly.
- Incubate overnight at 56 °C on a heated shaking block at a moderate speed.
- Next day, briefly spin tubes before opening.
- If samples are gingival tissues, continue to Step B12.
- If samples were collected with swabs, pull up the swab by grabbing it from the upper end with a pair of forceps, place a spin basket into the tube and now leave the swab into the basket.
- Take the tube with the basket and swab on top and proceed to briefly spin, to drain any liquid on the swab. Discard basket and swab.
- Add 200 µl of ethanol and mix well by vortexing.
- Briefly spin tubes before opening.
- Pipet the mixture into DNeasy mini spin column that was previously placed in a 2 ml collection tube (from now on all reagents are provided in the DNeasy Blood and Tissue Kit).
- Centrifuge at ≥ 6,000 x g for 1 min. Discard flow-through and collection tube.
- Place column in a new collection tube and add 500 µl of Buffer AW1.
- Centrifuge at ≥ 6,000 x g for 1 min. Discard flow-through and collection tube.
- Place column in a new collection tube and add 500 µl of Buffer AW2.
- Centrifuge at 20,000 x g for 3 min to dry column membrane. Discard flow-through and collection tube.
- Make sure column is dry before proceeding to next step, if it became wet, repeat Step B19 using a new collection tube.
- Place column in a new 1.5 ml Safe-lock Biopur tube (not included in the kit) and pipet 53 µl of Buffer AE directly onto the center of the membrane.
- Incubate for 5 min at room temperature.
- Centrifuge at ≥ 6,000 x g for 1 min.
- Measure DNA concentration in Spectrophotometer (Nanodrop, ND-1000).
- Clean work surface with 70% ethanol and then RNase AWAY reagent. UV-treat a plastic tube rack, pipettes, sterile forceps and spin baskets (consider one pair of forceps and one basket per sample).
- 16S rRNA gene amplification
DNA concentration from oral murine samples (in health/steady state) is low, particularly when using the oral swab procedure (typically around 2-3 ng/µl of total DNA). Therefore, we modified our protocols for the subsequent PCR reactions adding 4 µl per reaction regardless of their DNA concentration. We also adjusted amplification conditions by increasing the number of PCR cycles (from 25 to 35) and decreasing the annealing temperature (from 52.5 °C to 50 °C). Importantly, our non-template controls and water alone did not amplify with this modified protocol. All PCR reactions per sample are performed in duplicate, then PCR products are pooled before purification.- Clean work surface with 70% ethanol and then RNase AWAY reagent.
- Assemble master mix in a 2 ml LoBind tube, combine the required amounts of PCR water, dNTPs, MgSO4, Taq polymerase and buffer, according to the volume described below (per tube).
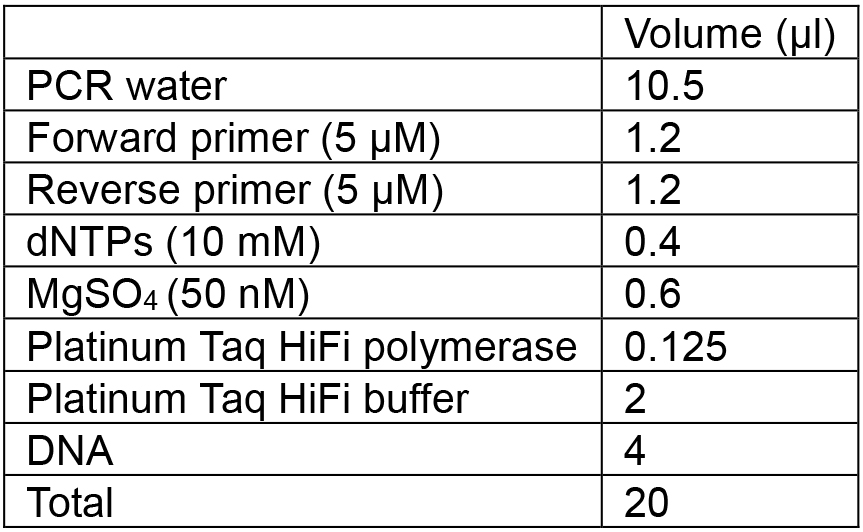
- Mix well by vortexing and add 13.6 µl of this mixture to each PCR tube.
- Add to each PCR tube the desired forward and reverse primer (both come with their own index sequence, for ‘dual indexing’).
- Add DNA template to each PCR tube.
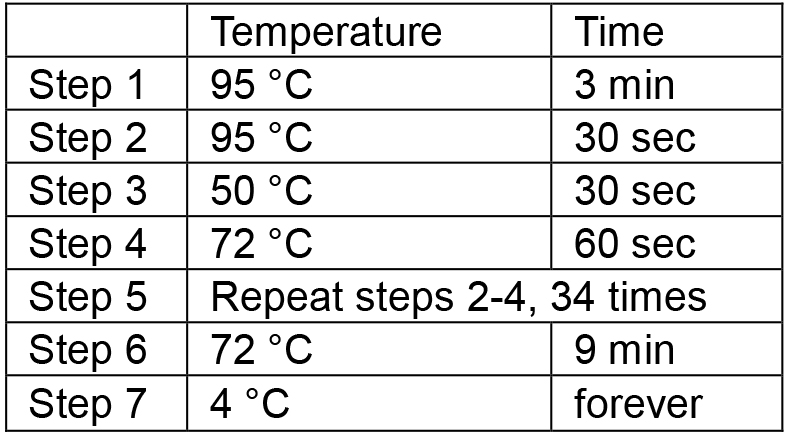
- Place the tubes in a thermal cycler and run the following PCR conditions:
- Merge the duplicated PCR products from one sample in a DNA low-bind 1.5 ml tube. Then, run a 1.2% agarose gel using a 100 bp ladder, stain it with ethidium bromide or the DNA dye of your preference and visualize the PCR products in a gel imager. The expected amplicon size is ~528 bp.
- Clean work surface with 70% ethanol and then RNase AWAY reagent.
- PCR product clean-up using the Agencourt AMPure XP system
- Clean work surface with 70% ethanol and then RNase AWAY reagent.
- Equilibrate AMPure XP Magnetic Particle Solution to room temperature (15-30 min).
- During that time prepare fresh 70% ethanol, aliquot the required amount of EB buffer (consider 40 µl per PCR product) and have ready a set of DNA low-bind 1.5 ml tubes.
- Measure the volume of each PCR product.
- Mix by vortexing the AMPure XP Magnetic Particle Solution, to re-suspend any magnetic particles that may have settled.
- According to the measured PCR product volume, add the appropriate amount of bead solution based on the following ratio (DNA:beads = 1:0.65).
- Vortex briefly and carefully, keeping the mixture in the bottom of the tubes. Incubate for 5 min at room temperature, for binding of DNA to beads.
- Place the tube onto a DynaMag-2 Magnet for 2 min or until beads separate from solution.
- While keeping the tubes in the magnet, carefully remove supernatant.
- Remove tubes from magnet and dispense 200 µl of 70% ethanol and pipet up and down to mix.
- Repeat Steps D8 and D9.
- Remove tubes from magnet and dispense 200 µl of 70% ethanol and pipet up and down to mix.
- Repeat Steps D8 and D9.
- While keeping the tubes in the magnet, open the tube lids and allow the beads to dry for approximately 5 min, making sure that the bead surface does not start to crack as this results in low DNA yield.
- Remove tubes from the magnet, add 40 µl of EB to each tube and mix well by pipetting.
- Place the tubes back in the magnet and wait for 2 min or until beads clearly separate from solution.
- Transfer ~35 µl of the supernatant to a new DNA low-bind 1.5 ml tube.
- Clean work surface with 70% ethanol and then RNase AWAY reagent.
- PCR product quantification using the Qubit dsDNA HS Assay Kit
- Equilibrate kit solutions to room temperature (15-30 min).
- Set up the required number of Qubit assay tubes for samples and standards (2 tubes for standards).
- Label the tube lids.
- Prepare working solution by diluting HS reagent 1:200 in HS buffer. Prepare the desired amount considering that each sample will require 198 µl and the standards 190 µl.
- Add 190 µl of working solution to the standard tubes and add 10 µl of each standard to the appropriate tube. Mix by vortexing 2-3 sec, avoiding bubble formation.
- Add 198 µl of working solution to the sample tubes and add 2 µl of each sample to the appropriate tube. Mix by vortexing 2-3 sec, avoiding bubble formation.
- Incubate for 2 min at room temperature in the dark.
- Calibrate Qubit Fluorometer with the standards and proceed to read samples.
Note: It is important to mention that the Qubit dsDNA HS Assay Kit detection range goes from 0.5 to 100 ng/µl. Samples below that detection range can be quantified, but their concentration may not be accurate.
- Equilibrate kit solutions to room temperature (15-30 min).
- PCR product quantification and visualization using the Experion DNA 1K Kit
We perform this step to confirm the purity of the PCR products after cleaning procedures. The size of the target DNA should correspond to a single peak at 528 bp.- Equilibrate kit reagents to room temperature (15-30 min).
- While waiting, make sure that the DNA concentration of the PCR products doesn’t exceed 50 ng/µl, if it is above this value dilute to 50 ng/µl. For our samples, DNA concentration is usually below this value.
- Vortex briefly and short spin all reagents.
- Prepare gel-stain solution (GS) by adding 12.5 µl of stain (ST) to gel tube (G).
- Vortex for 10 sec and transfer the GS solution to a spin-filter tube (provided in the kit).
- Centrifuge spin-filter tube at 2,400 x g for 15 min.
- Discard filter. Date the tube as this GS solution lasts for 1 month. This solution needs to be protected from light and stored at 4 °C.
- Prime the DNA chip by adding 9 µl of GS to priming well, then select C3 on priming station and press Start. Visually inspect for bubbles or incomplete priming.
- Pipet 9 µl of GS to other GS wells (3) in the DNA chip.
- Pipet 5 µl of loading buffer into L well and each sample well (11).
- Pipet 1 µl of DNA 1K ladder into well L.
- Pipet 1 µl of DNA into each sample well. Add 1 µl TE buffer or DNase-free water in wells that were not used.
- Place chip in the Experion vortex station and press mix.
- Run the chip in the Experion electrophoresis station.
- Equilibrate kit reagents to room temperature (15-30 min).
- PCR product pooling and sequencing
- Calculate the concentration in nM of each purified PCR product. Consider the amplicon size (528 bp) from Experion analyses and DNA concentration obtained from the clean PCR product using the Qubit.
- Dilute each PCR product to 4 nM using EB buffer. It is very important that PCR product concentrations and dilutions are accurate in order to obtain approximately equal number of reads per sample.
- Add 5 µl of each diluted PCR product to a 2.0 ml DNA low-bind tube, so samples are pooled in equimolar amounts. Measure library DNA concentration using Qubit assay as described before. Store at 4 °C.
- Pooled PCR products are ready to be sequenced on a MiSeq Instrument using the PhiX Control libraries and the MiSeq Reagent Kit v3 from Illumina, following the manufacturer’s standardized sequencing protocols.
- Prepare MiSeq reagents according to MiSeq® Reagent Kit v3 Reagent Preparation Guide.
- Considering DNA concentration measured in Step G4, proceed to denaturation and dilution steps described in MiSeq System Denature and Dilute Libraries Guide.
- For our samples, we diluted the library to 9 pM and combined it with 20 pM PhiX in a ratio that allows us to obtain a spike of PhiX of 14%.
- Calculate the concentration in nM of each purified PCR product. Consider the amplicon size (528 bp) from Experion analyses and DNA concentration obtained from the clean PCR product using the Qubit.
Data analysis
- Following sequencing, the data is processed using the software Mothur (Schloss et al., 2009). A detailed description of the analysis pipeline is available in our original article (Dutzan et al., 2017). Briefly, the initial steps of pre-processing are aimed at eliminating low-quality reads, assembling contigs and filtering according to size (200-400 bp). Subsequently, we follow the MiSeq SOP pipeline (https://www.mothur.org/wiki/MiSeq_SOP) as described in Kozich et al. (2013). After pre-processing, sequences are clustered into Operational Taxonomic Units (OTUs) (using a 97% similarity cutoff). For taxonomic classification, we use the Ribosomal Database Project classifier (Wang et al., 2007) adapted for Mothur, which allows classification of OTUs up to genus-level. To improve OTU taxonomical identification, we then obtain the representative sequence for each OTU and compare it against the NCBI 16S rRNA database using BLAST. Top matches (presenting at least 97% similarity and coverage) provide additional species-level taxonomy information for each OTU.
- For data visualization, we use the software R and R studio, in conjunction with the R packages ‘ggplot2’ and ‘RColorBrewer’.
- To get an overview of the taxonomical composition of the samples, we typically plot the relative abundance for the top OTUs (mean abundance > 1%) (Figure 3). Differences in relative abundance can be determined using appropriate statistical tests (considering if data are paired, follow a normal distribution, etc.) and adjusting for multiple comparisons. We often use the LDA effect Size (LEfSe) tool (Segata et al., 2011) to identify differentially represented OTUs, which is available at https://huttenhower.sph.harvard.edu/galaxy/.
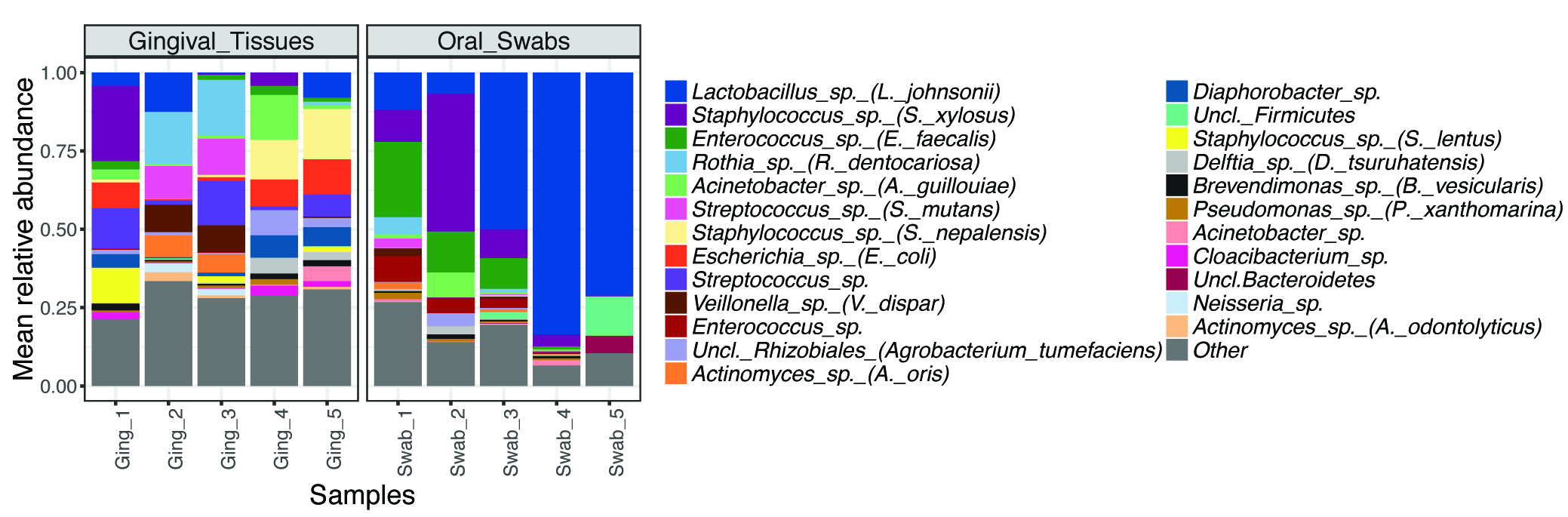
Figure 3. Most abundant OTUs in oral microbial communities from gingival tissues and oral mucosal surfaces. Example data from 10-week-old C57BL6 mice (n = 10). - To explore differences between communities (beta-diversity), samples can be compared using the ThetaYC distance, which measures dissimilarities in overall community structure. These data can be analyzed/visualized using Principal Coordinate Analysis (PCoA) (Figure 4).
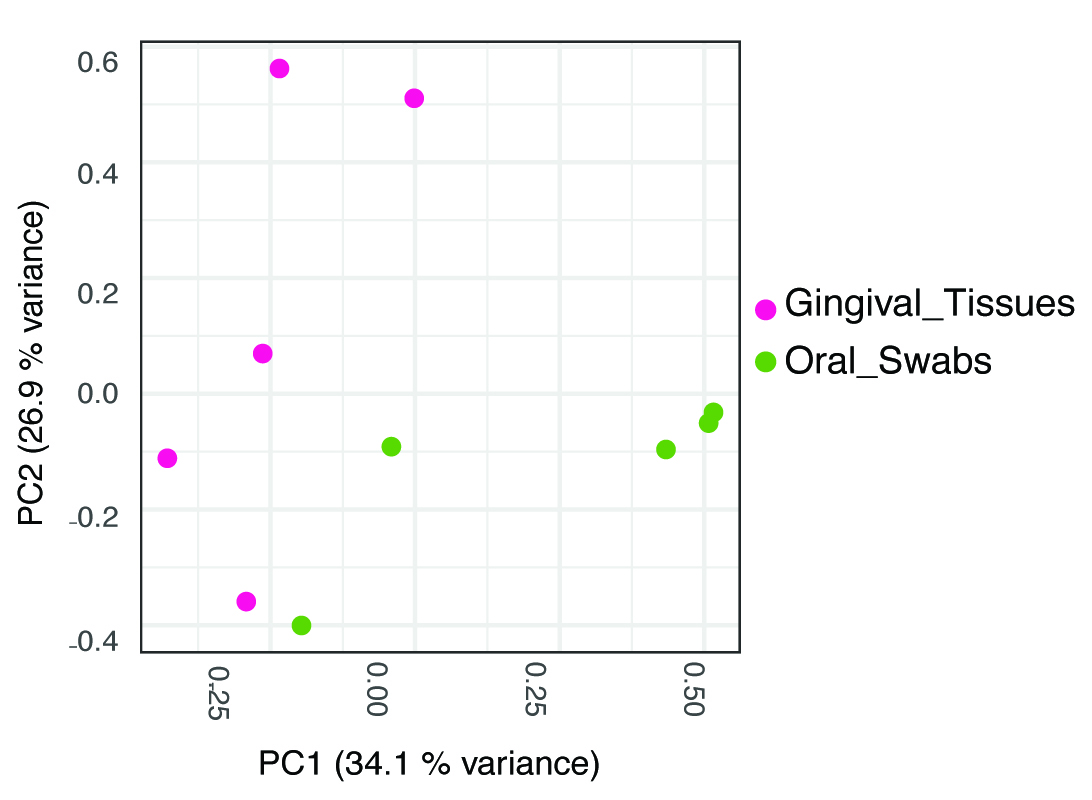
Figure 4. PCoA graph based on Theta YC distances. Samples from gingival and mucosal tissues cluster separately, indicating they harbor microbiomes with distinct global community structures (P < 0.001, determined by AMOVA).
Notes
Wear gloves, lab coat at all times and also consider wearing a mask when possible, to minimize risks of sample contamination during collection and processing.
Recipes
This protocol does not require recipes for reagents, almost all reagents come ready to use.
Acknowledgments
The authors thank Dr. Nicolas Dutzan for assistance with photography and Ms. Teresa Wild for carefully reviewing this manuscript. This work was funded in part by the Intramural Program of the National Institute of Dental and Craniofacial Research (NIDCR) (to N.M.M), by grants R01 DE021578 and R21DE023967 (to P.I.D) from NIDCR/NIH, by the BBSRC (BB/M025977/1 to J.E.K) and by the Manchester Collaborative Centre for Inflammation Research (to J.E.K). This protocol was adapted from methods previously published in Dutzan et al. (2017). The authors do not have any conflict of interest or competing interests to declare.
References
- Abusleme, L., Dupuy, A. K.,Dutzan, N., Silva, N., Burleson, J. A., Strausbaugh, L. D., Gamonal, J. andDiaz, P. I. (2013). The subgingival microbiome inhealth and periodontitis and its relationship with community biomass andinflammation. ISME J 7(5): 1016-1025.
- Abusleme, L. and Moutsopoulos, N.M. (2016). IL-17: overview and role in oralimmunity and microbiome. Oral Dis.
- Belkaid, Y. and Harrison, O. J.(2017). Homeostatic Immunity and theMicrobiota. Immunity 46(4): 562-576.
- Dutzan, N., Abusleme, L.,Bridgeman, H., Greenwell-Wild, T., Zangerle-Murray, T., Fife, M. E., Bouladoux,N., Linley, H., Brenchley, L., Wemyss, K., Calderon, G., Hong, B. Y., Break, T.J., Bowdish, D. M., Lionakis, M. S., Jones, S. A., Trinchieri, G., Diaz, P. I.,Belkaid, Y., Konkel, J. E. and Moutsopoulos, N. M. (2017). On-goingmechanical damage from mastication drives homeostatic Th17 cell responses atthe oral barrier. Immunity 46(1): 133-147.
- Dutzan, N., Abusleme, L., Konkel,J. E. and Moutsopoulos, N. M. (2016). Isolation, characterization andfunctional examination of the gingival immune cell network. J Vis Exp(108): 53736.
- Fadrosh, D. W., Ma, B., Gajer, P.,Sengamalay, N., Ott, S., Brotman, R. M. and Ravel, J. (2014). Animproved dual-indexing approach for multiplexed 16S rRNA gene sequencing on theIllumina MiSeq platform. Microbiome 2(1): 6.
- Griffen, A. L., Beall, C. J.,Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., Podar, M. andLeys, E. J. (2012). Distinct and complex bacterialprofiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6(6): 1176-1185.
- Human Microbiome Project, C. (2012). Structure,function and diversity of the healthy human microbiome. Nature 486: 207-214.
- Ivanov, II, Atarashi, K., Manel,N., Brodie, E. L., Shima, T., Karaoz, U., Wei, D., Goldfarb, K. C., Santee, C.A., Lynch, S. V., Tanoue, T., Imaoka, A., Itoh, K., Takeda, K., Umesaki, Y.,Honda, K. and Littman, D. R. (2009). Induction of intestinal Th17 cellsby segmented filamentous bacteria. Cell 139(3): 485-498.
- Kostic, A. D., Howitt, M. R. andGarrett, W. S. (2013). Exploring host-microbiotainteractions in animal models and humans. GenesDev 27(7): 701-718.
- Kozich, J.J., Westcott, S. L., Baxter, N. T., Highlander, S. K. and Schloss, P. D.(2013). Development of a dual-indexsequencing strategy and curation pipeline for analyzing amplicon sequence dataon the MiSeq Illumina sequencing platform. ApplEnviron Microbiol 79(17): 5112-5120.
- Moutsopoulos, N. M., Chalmers, N.I., Barb, J. J., Abusleme, L., Greenwell-Wild, T., Dutzan, N., Paster, B. J.,Munson, P. J., Fine, D. H., Uzel, G. and Holland, S. M. (2015). Subgingivalmicrobial communities in Leukocyte Adhesion Deficiency and their relationshipwith local immunopathology. PLoS Pathog 11(3):e1004698.
- Naik, S., Bouladoux, N., Wilhelm,C., Molloy, M. J., Salcedo, R., Kastenmuller, W., Deming, C., Quinones, M.,Koo, L., Conlan, S., Spencer, S., Hall, J. A., Dzutsev, A., Kong, H., Campbell,D. J., Trinchieri, G., Segre, J. A. and Belkaid, Y. (2012). Compartmentalizedcontrol of skin immunity by resident commensals. Science 337(6098): 1115-1119.
- RCore Team (2017). R:A language and environment for statistical computing. RFoundation for Statistical Computing. Vienna, Austria.
- RStudioTeam (2015). RStudio:Integrated Development for R. RStudio. Inc.,Boston, MA.
- Schloss, P. D., Westcott, S. L.,Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A.,Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B.,Thallinger, G. G., Van Horn, D. J. and Weber, C. F. (2009). Introducingmothur: open-source, platform-independent, community-supported software fordescribing and comparing microbial communities. Appl Environ Microbiol 75(23): 7537-7541.
- Segata, N., Izard, J., Waldron,L., Gevers, D., Miropolsky, L., Garrett, W. S. and Huttenhower, C. (2011). Metagenomicbiomarker discovery and explanation. GenomeBiol 12(6): R60.
- Sundquist, A., Bigdeli, S.,Jalili, R., Druzin, M. L., Waller, S., Pullen, K. M., El-Sayed, Y. Y., Taslimi,M. M., Batzoglou, S. and Ronaghi, M. (2007). Bacterial flora-typing withtargeted, chip-based Pyrosequencing. BMCMicrobiol 7: 108.
- Turnbaugh, P. J., Ley, R. E.,Mahowald, M. A., Magrini, V., Mardis, E. R. and Gordon, J. I. (2006). Anobesity-associated gut microbiome with increased capacity for energy harvest. Nature 444(7122): 1027-1031.
- Wang, Q., Garrity, G. M., Tiedje,J. M. and Cole, J. R. (2007). Naive Bayesian classifier forrapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16): 5261-5267.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Abusleme, L., Hong, B., Hoare, A., Konkel, J. E., Diaz, P. I. and Moutsopoulos, N. M. (2017). Oral Microbiome Characterization in Murine Models. Bio-protocol 7(24): e2655. DOI: 10.21769/BioProtoc.2655.
Category
Microbiology > Microbe-host interactions > In vivo model > Mammal
Immunology > Mucosal immunology > Digestive tract
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









