- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ex vivo Trophoblast-specific Genetic Manipulation Using Lentiviral Delivery
Published: Vol 7, Iss 24, Dec 20, 2017 DOI: 10.21769/BioProtoc.2652 Views: 8145
Reviewed by: Giusy TornilloIrit AdiniAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vivo Electroporation of Skeletal Muscle Fibers in Mice
Steven J. Foltz [...] Hyojung J. Choo
Jul 5, 2023 1823 Views

Cochlear Organ Dissection, Immunostaining, and Confocal Imaging in Mice
Chenyu Chen [...] Dongdong Ren
Jan 20, 2025 3759 Views

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2627 Views
Abstract
In this protocol report, we describe a lentiviral gene delivery technique for genetic modification of the rat trophoblast cell lineage. Lentiviral packaged gene constructs can be efficiently and specifically delivered to the trophoblast cell lineage of the blastocyst. The consequences of ‘gain-of-function’ and ‘loss-of-function’ blastocyst manipulations can be evaluated with in vitro outgrowth assays or following transfer to pseudopregnant rats.
Keywords: BlastocystBackground
The placenta functions as a conduit between the mother and the developing fetus and is essential for viviparity (Georgiades et al., 2002). It is specialized to facilitate and coordinate maternal adaptations to pregnancy and fetal development (Soares et al., 2014; Burton et al., 2016). The placenta contains trophoblast cells, which perform several specialized functions. Acquisition of trophoblast cell specializations requires highly regulated differentiation of trophoblast stem and progenitor cell populations (Maltepe and Fisher, 2015). The mature rat placenta is comprised of two morphologically and functionally distinct compartments: the junctional zone and the labyrinth zone (Soares et al., 2012). Progenitor cells, spongiotrophoblast cells, glycogen trophoblast cells, and polyploid trophoblast giant cells comprise the junctional zone. An invasive trophoblast lineage arises from progenitors within the junctional zone region (Ain et al., 2003; Soares et al., 2014). During the last week of gestation, these cells move out of the placenta and invade into the maternal uterine mesometrial compartment (Ain et al., 2003; Pijnenborg and Vercruysse, 2010). The innermost layer of the placenta (proximal to the developing fetus) is called the labyrinth zone, which consists of trophoblast cells and fetal vasculature derived from the allantois. Progenitor trophoblast cells within the labyrinth zone fuse to form syncytia, which provide barriers between maternal and fetal compartments (Soares et al., 2012). The labyrinth zone consists of an elaborate branched structure, providing a large surface area for nutrient, waste and gas exchange with the fetus (Knipp et al., 1999; Watson and Cross, 2005).
Specific modifications of the trophoblast lineage can be achieved using lentiviral transduction of the outer layer of the embryo at the blastocyst stage, termed trophectoderm (Georgiades et al., 2007; Malashicheva et al., 2007; Okada et al., 2007; Lee et al., 2009) (Figure 1). This method allows for efficient manipulation of all trophoblast cell lineages. It also facilitates discrimination between trophoblast and embryonic contributions to phenotypic outcomes arising from global genetic manipulations. The experimental basis for selective trophoblast viral infection is directly related to the structure of the blastocyst and the tight epithelium formed by the outer trophectoderm, which effectively restricts viral particle access to the inner cell mass (Georgiades et al., 2007; Malashicheva et al., 2007; Okada et al., 2007; Lee et al., 2009). This method can be utilized to investigate the consequences of both ‘gain-of-function’ and ‘loss-of-function’ manipulations of the trophoblast lineage, which can provide insights into molecular mechanisms controlling placental development (Georgiades et al., 2007; Malashicheva et al., 2007; Okada et al., 2007; Lee et al., 2009; Morioka et al., 2009; Kent et al., 2011; Chakraborty et al., 2016; Muto et al., 2016).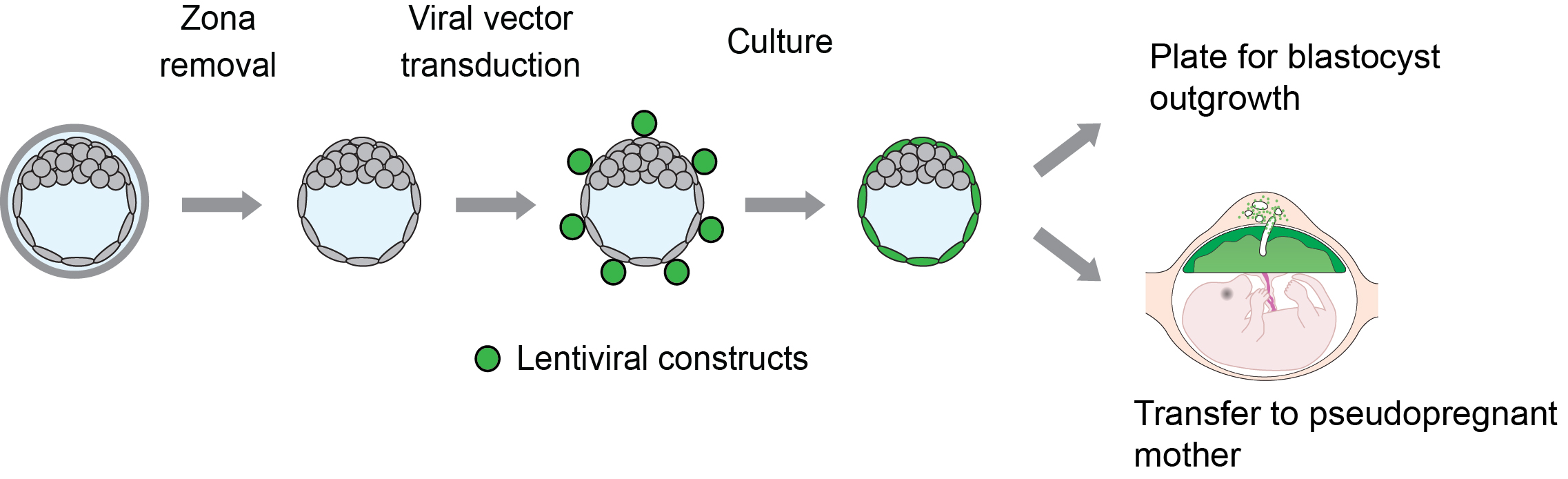
Figure 1. Schematic showing experimental plan for lentiviral transduction of blastocysts and downstream analyses. Blastocysts are transduced with lentiviral particles (shown in green) expressing specific ‘gain-of-function’ or ‘loss-of-function’ constructs and cultured for 72 h for outgrowth assays or directly transferred to day 3.5 pseudopregnant animals for in vivo analyses.
Materials and Reagents
- Multi-well glass agglutination plates (Scientific Device Laboratory, catalog number: 074-3032B )
- Tissue culture plate, 6-well (MIDSCI, catalog number: TP92006 )
- Needle,18 gauge (BD, BD Biosciences, catalog number: 305195 )
- Syringe, 1 ml
- Tissue culture plate, 12-well (MIDSCI, catalog number: TP92012 )
- Glass mouth pipette (Drummond Scientific, catalog number: 2-000-100 )
- Aspirator tube assembly, 15” (Drummond Scientific, catalog number: 2-000-000 )
- Tissue culture plate, 4-well (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 176740 )
- Plastic non-tissue culture Petri plates (Fisher Scientific, catalog number: AS4052 )
- Female rats (8-10 weeks of age)
- Lentiviral packaging reagents: pRSV-Rev (Addgene, catalog number: 12253 ), pMDLg/pRRE (Addgene, catalog number: 12251 ), VSVG envelope plasmid (pMD2.G, Addgene, catalog number: 12259 )
- Sterile saline solution (0.9% Sodium Chloride)
- Opti-MEM culture medium (Thermo Fisher Scientific, catalog number: 51985034 )
- Lipofectamine 2000 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668-027 )
- p24 enzyme-linked immunoassay kit (Takara Bio, Clontech, catalog number: 632200 )
- M2 medium (Millipore Sigma, catalog number: MR-015-D )
- KSOM medium (Millipore Sigma, catalog number: MR-121-D )
- Acid Tyrode’s solution (Sigma-Aldrich, catalog number: T1788 )
- Mineral oil (Sigma-Aldrich, catalog number: M8410 )
- 4% paraformaldehyde (Sigma-Aldrich, catalog number: 158127 )
- RPMI 1640 (Thermo Fisher Scientific, GibcoTM, catalog number: 11875093 )
- Fetal bovine serum, heat inactivated (Sigma-Aldrich, catalog number: F2442 )
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- Sodium pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 11360070 )
- Penicillin, and streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- (Optional) Clontech LentiX titration kit (Takara Bio, Clontech, catalog number: 631235 )
- Outgrowth culture medium (see Recipes)
Equipment
- Centrifuge (Beckman Coulter, model: OptimaTM L-100XP , catalog number: 392050)
- Standard inverted light microscope or stereo microscope
- Standard CO2 cell culture incubator, 5% CO2, 37 °C
Software
- ImageJ software (NIH)
Procedure
- Preparation of rats for timed pregnancy
- Animals are maintained in an environmentally-controlled facility with lights on from 06:00 to 20:00 (14 h light:10 h dark cycle) and are allowed free access to food and water.
- Female rats (8-10 weeks of age) are placed with male rats (> 3 months of age) of the same strain.
- Confirmation of mating is determined by inspection of vaginal lavages. Plastic pipettes are loaded with sterile saline solution. Saline is delivered to the vagina. Vaginal lavages are transferred to clean multi-well glass plates and observed under a microscope. The presence of sperm in the vaginal lavage is considered gestation day (gd) 0.5.
- In the rat, vaginal retention of seminal plugs is not common. Thus, it is necessary to obtain vaginal lavages as opposed to visual identification of seminal plugs.
- Lavages should be obtained between 08:00-10:00 for confirmation of sperm. If collected later, sperm may not be easily visible in the vaginal lavage and may lead to a false negative.
- Plastic pipettes should be washed thoroughly before obtaining lavages, so that there is no sperm contamination from one animal to the next, which may lead to a false positive.
- Animals are maintained in an environmentally-controlled facility with lights on from 06:00 to 20:00 (14 h light:10 h dark cycle) and are allowed free access to food and water.
- Virus production and concentration
- 293FT cells are split into 6-well plates at 70-80% confluency.
- After 16-20 h post plating, the medium is changed to Opti-MEM medium at 1.5 ml per well.
- Proceed to transfection 1 h following the medium change.
- Lentivirus is produced following transient transfection of a transducing vector and third generation packaging system plasmids. Prepare transfection mix utilizing Lipofectamine 2000 following manufacturer’s protocol. Add 500 ng of the transducing vector (shRNA construct or overexpression construct), 200 ng of pRSV-Rev (Addgene plasmid 12253), 500 ng of pMDLg/pRRE (Addgene plasmid 12251) and 300 ng of VSVG envelope plasmid (pMD2.G, Addgene plasmid 12259).
- Transfection mix is removed after 8 h, and 2 ml of Opti-MEM medium supplemented with 5% fetal bovine serum is added.
- The medium is removed after 20 h and collected in tubes stored at 4 °C. Two collections are performed in a period of 40-48 h.
- Centrifuge the culture supernatant to remove cell debris, filter sterilize and concentrate by ultracentrifugation (35,000 x g for 3 h). Store at -80 °C until used for transduction. Do not freeze-thaw.
- Lentiviral vector titers should be determined by measurement of p24 gag antigen by enzyme-linked immunoassay.
- 293FT cells are split into 6-well plates at 70-80% confluency.
- Embryo collection
- Reproductive tracts of gd 4.5 pregnant rats are collected between 08:30 to 09:30.
- Uteri are dissected from adipose tissue and removed by cutting at the utero-tubal junction and cervix.
- Uteri are flushed with M2 medium from the cervical end with an 18-gauge needle connected to a 1 ml syringe.
- The flushed exudate is collected in a well of a 12-well tissue culture plate.
- Blastocysts are retrieved through a mouth pipette and transferred to a droplet of KSOM medium.
- Blastocysts are maintained at 37 °C in a CO2 incubator for 30 min to stabilize from the handling-associated stress.
- The timing of collection is very important for obtaining blastocyst stage embryos. If collected too early, the embryos will be in the oviduct and no blastocyst stage embryos will be obtained by flushing the uterus. If the collection is done too late, the embryos begin to attach to the uterus and it becomes very difficult to retrieve them by flushing the uterus.
- Embryos should be immediately transferred to KSOM medium at 37 °C in a 5% CO2 incubator and not left in unbuffered medium for extended periods.
- For details regarding embryo flushing and collection, please refer to Chiu et al., 2010.
- Reproductive tracts of gd 4.5 pregnant rats are collected between 08:30 to 09:30.
- Removal of zona pellucida from blastocysts
- Place two separate droplets of Acid Tyrode’s solution (approximately 50 µl droplet size) in a Petri dish.
- In a second Petri dish, prepare 5 individual droplets of KSOM medium (approximately 40 µl droplet size).
- Under an inverted microscope, blastocysts are transferred to the first Acid Tyrode’s solution droplet and zona pellucida integrity monitored. If the zona pellucida dissolution is incomplete, then pipette the blastocysts into the second droplet and then immediately transfer them to the KSOM medium droplets.
- After successful zona pellucida removal and 5 KSOM washes, transfer the blastocysts into KSOM droplet covered with mineral oil. Mineral oil should completely cover the KSOM droplet and the total volume of mineral oil necessary will depend on the Petri dish size (2-3 ml will be sufficient to cover the KSOM droplet).
- Incubate the blastocysts at 37 °C for further stabilization.
- The survival of the embryo depends on the incubation time and exposure to Acid Tyrode’s solution. Overexposure to Acid Tyrode’s solution leads to disrupted and collapsed embryos, which will not be suitable for outgrowth or in vivo transfer experiments.
- When embryos are transferred to Acid Tyrode’s solution, constant observation is necessary to remove them immediately when the zona pellucida begins to dissociate. Once the zona pellucida is removed, the embryos should be immediately washed and transferred to a KSOM droplet.
- After removal of zona pellucida, embryos become sticky and tend to aggregate. Careful and gentle mouth pipetting is recommended to detach the embryos and proceed with further wash steps.
- Place two separate droplets of Acid Tyrode’s solution (approximately 50 µl droplet size) in a Petri dish.
- Virus incubation and washes
- Lentiviral particles containing ‘gain-of-function’ or ‘loss-of-function’ constructs are generated, concentrated and stored, as previously reported (Lee et al., 2009).
- Add 10 µl of concentrated virus in a 30 µl KSOM droplet.
- Transfer blastocysts into the KSOM droplet containing virus particles. The droplet should be covered with mineral oil (2-3 ml will be sufficient to cover the KSOM droplet).
- Incubate the blastocysts in virus at 37 °C for 4 h.
- After the incubation, wash the blastocysts sequentially in 10-KSOM droplets.
- After the 10th wash, transfer the blastocysts into a droplet of KSOM covered with mineral oil and incubate at 37 °C for 15 min.
- The p24 coat protein measurement gives a relative assessment of virus concentration and does not necessarily indicate efficacy of infectivity of the virus batch.
- Alternative qPCR approaches (Clontech LentiX titration kit) can also be utilized to measure virus titers. 1 x 108 through 3 x 108 infectious unit/ml can be used to incubate blastocysts for effective transduction.
- Virus concentrations of 500 ng of p24/ml have been effective for transduction. However, effective viral particle concentrations need to be empirically determined for each experimental application.
- Lentiviral particles containing ‘gain-of-function’ or ‘loss-of-function’ constructs are generated, concentrated and stored, as previously reported (Lee et al., 2009).
- Preparation for blastocyst outgrowth
- In a single well of a 4-well tissue culture plate, add 750 µl of outgrowth culture medium (see Recipes).
- Add a single virally-manipulated blastocyst to each well and incubate at 37 °C.
- Observe the blastocysts daily, until the blastocysts have attached to the 4-well tissue culture plate.
- Once the blastocysts are attached, remove the medium and add fresh medium. Usually, it takes 2-3 days for the blastocysts to hatch and attach.
- On day 5 of incubation, outgrowth will be observed in control blastocysts (Figure 2).
- Outgrowths can be fixed in 4% paraformaldehyde for immunofluorescence, or can be harvested for further biochemical/molecular analysis.
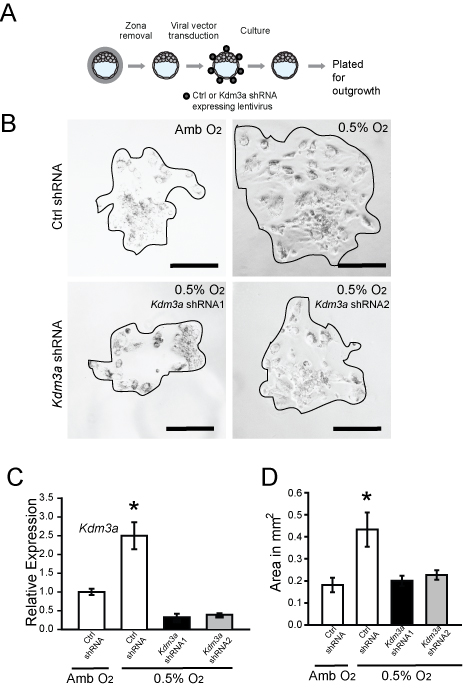
Figure 2. Effects of oxygen tension and KDM3A expression on blastocyst outgrowth. A. Schematic showing experimental plan for lentiviral transduction of blastocysts and outgrowth assay. Blastocysts were transduced with control (Ctrl) or Kdm3a shRNA and cultured for 72 h to allow hatching from the zona pellucida. The attached blastocysts were exposed to ambient (Amb) or low oxygen (0.5% O2) for 24 h and analyzed. B. Representative images of blastocyst outgrowths from Ctrl shRNA and exposed to Amb, Ctrl shRNA and exposed to 0.5% O2, and Kdm3a shRNAs and exposed to 0.5% O2. C. Measurement of Kdm3a transcripts in control and knockdown cultures was measured by qRT-PCR. Asterisks indicate significant differences among groups (n = 6/group; *P < 0.05). D. The bar graph shows quantification of outgrowth area in square millimeters. The area of the outgrowth was measured using ImageJ software (Ctrl shRNA + Amb, n = 6; Ctrl shRNA + 0.5% O2, Kdm3a shRNA1 + 0.5% O2, n = 10; Kdm3a shRNA2 + 0.5% O2, n = 10; *P < 0.05). Data presented in C and D were analyzed with ANOVA and Student-Newman-Keuls test. This figure appeared in Chakraborty et al. (2016).
- In a single well of a 4-well tissue culture plate, add 750 µl of outgrowth culture medium (see Recipes).
- In vivo transfer of virally-manipulated blastocysts to pseudopregnant rats
- Pseudopregnant rats are prepared by mating cycling female rats to vasectomized male rats in wire bottom cages. Mating is verified by the identification of seminal plugs beneath the cage. Detection of the seminal plugs is enhanced by placing black paper beneath the wire bottom cages. Detection of seminal plugs is considered day 0.5 of pseudopregnancy.
- Following the 4-h incubation of blastocysts with lentiviral vectors and subsequent washes (as described above), then the virally-manipulated blastocysts are transferred to uteri of day 3.5 pseudopregnant rats (~8 blastocysts per uterine horn).
- Pregnancies can then be terminated at desired times during gestation and placentation sites interrogated using histological and biochemical approaches (Ain et al., 2006).
- Pseudopregnant rats are prepared by mating cycling female rats to vasectomized male rats in wire bottom cages. Mating is verified by the identification of seminal plugs beneath the cage. Detection of the seminal plugs is enhanced by placing black paper beneath the wire bottom cages. Detection of seminal plugs is considered day 0.5 of pseudopregnancy.
Data analysis
For data processing and analyses, please refer to Chakraborty et al. (2016).
Recipes
- Outgrowth culture medium
RPMI 1640
20% fetal bovine serum
100 µM 2-mercaptoethanol
1 mM sodium pyruvate
50 µM penicillin, and 50 U/ml streptomycin
Acknowledgments
This protocol was adapted from Lee et al. (2009) and Chakraborty et al. (2016). Funding for this work was provided by the NIH, HD020676 and HD079363. Authors declare no conflict of interest or competing interests.
References
- Ain, R., Canham, L. N. and Soares, M. J. (2003). Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: novel endocrine phenotype and regulation. Dev Biol 260(1): 176-190.
- Ain, R., Konno, T., Canham, L. N. and Soares, M. J. (2006). Phenotypic analysis of the rat placenta. Methods Mol Med 121: 295-313.
- Burton, G. J., Fowden, A. L. and Thornburg, K. L. (2016). Placental origins of chronic disease. Physiol Rev 96(4): 1509-1565.
- Chakraborty, D., Cui, W., Rosario, G. X., Scott, R. L., Dhakal, P., Renaud, S. J., Tachibana, M., Rumi, M. A., Mason, C. W., Krieg, A. J. and Soares, M. J. (2016). HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc Natl Acad Sci U S A 113(46): E7212-E7221.
- Chiu, S. Y., Maruyama, E. O. and Hsu, W. (2010). Derivation of mouse trophoblast stem cells from blastocysts. J Vis Exp 8(40).
- Georgiades, P., Cox, B., Gertsenstein, M., Chawengsaksophak, K. and Rossant, J. (2007). Trophoblast-specific gene manipulation using lentivirus-based vectors. Biotechniques 42(3): 317-318, 320, 322-325.
- Georgiades, P., Ferguson-Smith, A. C. and Burton, G. J. (2002). Comparative developmental anatomy of the murine and human definitive placentae. Placenta 23(1): 3-19.
- Kent, L. N., Rumi, M. A., Kubota, K., Lee, D. S. and Soares, M. J. (2011). FOSL1 is integral to establishing the maternal-fetal interface. Mol Cell Biol 31(23): 4801-4813.
- Knipp, G. T., Audus, K. L. and Soares, M. J. (1999). Nutrient transport across the placenta. Adv Drug Deliv Rev 38(1): 41-58.
- Lee, D. S., Rumi, M. A., Konno, T. and Soares, M. J. (2009). In vivo genetic manipulation of the rat trophoblast cell lineage using lentiviral vector delivery. Genesis 47(7): 433-439.
- Malashicheva, A., Kanzler, B., Tolkunova, E., Trono, D. and Tomilin, A. (2007). Lentivirus as a tool for lineage-specific gene manipulations. Genesis 45(7): 456-459.
- Maltepe, E. and Fisher, S. J. (2015). Placenta: the forgotten organ. Annu Rev Cell Dev Biol 31: 523-552.
- Morioka, Y., Isotani, A., Oshima, R. G., Okabe, M. and Ikawa, M. (2009). Placenta-specific gene activation and inactivation using integrase-defective lentiviral vectors with the Cre/LoxP system. Genesis 47(12): 793-798.
- Muto, M., Fujihara, Y., Tobita, T., Kiyozumi, D. and Ikawa, M. (2016). Lentiviral vector-mediated complementation restored fetal viability but not placental hyperplasia in Plac1- deficient mice. Biol Reprod 94: 6.
- Okada, Y., Ueshin, Y., Isotani, A., Saito-Fujita, T., Nakashima, H., Kimura, K., Mizoguchi, A., Oh-Hora, M., Mori, Y., Ogata, M., Oshima, R. G., Okabe, M. and Ikawa, M. (2007). Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat Biotechnol 25(2): 233-237.
- Pijnenborg, R. and Vercruysse, L. (2010). Chap. 13: Animal models of deep trophoblast invasion. In: Pijnenborg, R., Brosens, I. and Romero, R. (Eds.). Placental bed disorders: basic science and its translation to obstetrics. Cambridge University Press pp: 127-139.
- Soares, M. J., Chakraborty, D., Karim Rumi, M. A., Konno, T. and Renaud, S. J. (2012). Rat placentation: an experimental model for investigating the hemochorial maternal-fetal interface. Placenta 33(4): 233-243.
- Soares, M. J., Chakraborty, D., Kubota, K., Renaud, S. J. and Rumi, M. A. (2014). Adaptive mechanisms controlling uterine spiral artery remodeling during the establishment of pregnancy. Int J Dev Biol 58(2-4): 247-259.
- Watson, E. D. and Cross, J. C. (2005). Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 20: 180-193.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chakraborty, D., Muto, M. and Soares, M. J. (2017). Ex vivo Trophoblast-specific Genetic Manipulation Using Lentiviral Delivery. Bio-protocol 7(24): e2652. DOI: 10.21769/BioProtoc.2652.
Category
Developmental Biology > Cell growth and fate > Blastocyst
Cell Biology > Cell engineering > Lentiviral delivery
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









