- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Drosophila Model of Leishmania amazonensis Infection
Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2640 Views: 7624
Reviewed by: Adler R. DillmanPascale GeirardAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Infection of Drosophila Primary Hemocytes
Charles Tracy and Helmut Krämer
Jun 5, 2017 11196 Views

Hypochlorous Acid Staining with R19-S in the Drosophila Intestine upon Ingestion of Opportunistic Bacteria
Salma Hachfi [...] Armel Gallet
May 20, 2019 6448 Views
Abstract
This protocol describes how to generate and harvest antibody-free L. amazonensis amastigotes, and how to infect adult Drosophila melanogaster with these parasites. This model recapitulates key aspects of the interactions between Leishmania amastigotes and animal phagocytes.
Keywords: Drosophila melanogasterBackground
Leishmaniasis, caused by Leishmania protozoans, affects the skin, mucosa or internal organs, depending on the parasite species and immunological status of the host. Although the mouse model of infection has provided most of our understanding about this parasitic infection, it is not well suited for large-scale exploratory approaches. In our lab, we exploited the tractable and powerful genetics of the fruit fly Drosophila melanogaster to establish a model of Leishmania infection and perform a small-scale screen to identify host genes involved in the phagocytosis of these parasites (Okuda et al., 2016). In particular, a set of genes possibly linked to phagocytosis was specifically knocked-down in the fly hemocytes, using the UAS-GAL4 expression systems (Brand and Perrimon, 1993) and the survival and parasite burden of mutant-infected flies were monitored to identify factors modulating the infection.
Materials and Reagents
- 15 ml conical tube (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339650 )
- Cell culture flasks (T-flask) 80 cm2 (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 178905 )
- T-flask 175 cm2 (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 178883 )
- Cell scraper (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 179707 )
- 10 ml serological pipette (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 170356 )
- 1.5 ml microcentrifuge tube (Corning, Costar®, catalog number: 3622 )
- 96-well plate (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 167008 )
- Pipette tips (Fisher Scientific, catalog numbers: 21-402-269 , 21-402-569 , 21-402-589 )
Manufacturer: Thermo Fisher Scientific, catalog numbers: 3752RI , 3782RI . - Disposable counting chamber (Bio-Rad Laboratories, catalog number: 1450003 )
- Pellet pestles (Fisher Scientific, catalog number: 12-141-364 )
- Fly vials (Genesee Scientific, Flystuff, catalog number: 32-116SB )
- Leishmania amazonensis (strain RAT/BA/LV78, Dutta et al., 2005)
- BALB/c bone marrow derived macrophages (BMDM)
Note: Material and reagents are described in the Bio-protocol paper of Chen, 2013. Balb/c mice were obtained from The Jackson Laboratories. The femurs and tibias of one mouse generate approximately 3 x 108 macrophages. - Phosphate-buffered saline (PBS) (Corning, catalog number: 21-031-CV )
- RPMI 1640 (Corning, catalog number: 10-040-CV )
- Percoll (GE Healthcare, catalog number: 17-0891-01 )
- Bromophenol blue (Fisher Scientific, catalog number: BP115-25 )
- Fly food (FlyStuff Nutri-Fly Molasses Formulation or Nutri-FlyTM Bloomington Formulation, Genesee Scientific, Flystuff, catalog numbers: 66-116 )
- Yellow Cornmeal Fly food (Genesee Scientific, Flystuff, catalog number: 62-101 )
- Schneider’s insect medium (Thermo Fisher Scientific, GibcoTM, catalog number: 21720024 )
- Penicillin/streptomycin (Corning, catalog number: 30-002-Cl )
- Tetracyclin (Sigma-Aldrich, catalog number: T7660 )
- Tunicamycin (Cayman Chemical, catalog number: 11445 )
- 70% ethanol
- 199 media (Corning, catalog number: 90-050-PB )
- Heat inactivated fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S12450 )
- HEPES (Sigma-Aldrich, catalog number: H3784-100G )
- Sodium succinate (Sigma-Aldrich, catalog number: S9637 )
- Hemin (Sigma-Aldrich, catalog number: H9039 )
- Tryptic soy broth (BD, catalog number: 211825 )
- 199 media to grow promastigotes (see Recipes)
- 199 media for promastigote to axenic amastigote differentiation (see Recipes)
Equipment
- Centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: SorvallTM LegendTM RT ), equipped with swinging buckets for 15 ml conical tubes
- Incubator (VWR® Water Jacketed CO2 Incubators)
- Glass dounce with grinding chamber and Teflon rod, clearance of 0.004” to 0.006” (DWK Life Sciences, Wheaton, catalog number: 358011 )
- Hemocytometer Hausser Scientific Bright-LineTM Counting Chamber (Hausser Scientific, catalog number: 3110 )
- Capillary puller (NARISHIGE, model: PC-100 )
- Nanoliter injector (Drummond Scientific, model: Nanoject II )
- Drummond Scientific Capillaries for Nanojet II (Pipette.com, catalog number: 3-000-203-G/X)
Manufacturer: Drummond Scientific, catalog number: 3-000-203-G/X . - Electronic caliper (Mitutoyo, catalog number: 500-197-30 )
- CO2 pad (Genesee Scientific, FlyStuff, catalog number: 59-114 )
- Stereomicroscope (Nikon Instruments, model: SMZ645 )
- Multichannel pipette (Thermo Fisher Scientific, Thermo ScientificTM, model: FinnpipetteTM F2 )
- Pellet grinder (Fisher Scientific, catalog number: 12-141-361 )
- Fine tweezers (Fisher Scientific, catalog number: 12-000-122 )
- Fluorescence microscope (Nikon Instruments, model: Eclipse E400 ) equipped with an imaging system (camera: Nikon Instruments, model: Ds-Qi1 , imaging software: NIS-Elements)
Software
- Imaging software: NIS-Elements Basic Reseach, Microscope Imaging Software (Nikon Instruments)
- GraphPad Prism7 (GraphPad Software)
- CellProfiler cell image analysis software (www.cellprofiler.org)
Procedure
- Leishmania culture, to generate amastigotes
- Thaw a vial with promastigotes and cultivate in 10 ml of 199 medium (see Recipes) supplemented with 10% of fetal bovine serum and 3 µg/ml of tunicamycin (selection marker for DsRed expression). Start a 1 x 106 parasites/ml culture in 80 cm2 T-flasks for 3 days at 26 °C. Parasites actively proliferate present > 90% viability on the third day.
- Centrifuge the culture 700 x g for 10 min in a 15 ml conical tube.
- Remove the supernatant and resuspend the parasites in 199 media (see Recipe 2) at a final concentration of 107 parasites/ml. Transfer the parasites to a T-flask.
- Incubate the culture at 34 °C for 3-4 days to allow promastigote to amastigote differentiation. Parasites with amastigote morphology (oval shape and no extracellular flagellum) are seen as early as 1 day after differentiation induction. However, the parasites require 3 days to complete differentiation.
Note: Axenic amastigotes are commonly used in research, but they are not identical to intracellular amastigotes (Gupta et al., 2001). - Centrifuge at 700 x g for 10 min. Wash with 10 ml PBS. Repeat this wash once.
- Transfer 1 x 108 parasites to a culture of Balb/c BMDM cultivated in 175 cm2 T-flasks (2 x 107 BMDM/flask).
- Keep the cultures at 34 °C and feed the cells with fresh RPMI 1640 supplemented with 10% FBS every 5 days.
- Thaw a vial with promastigotes and cultivate in 10 ml of 199 medium (see Recipes) supplemented with 10% of fetal bovine serum and 3 µg/ml of tunicamycin (selection marker for DsRed expression). Start a 1 x 106 parasites/ml culture in 80 cm2 T-flasks for 3 days at 26 °C. Parasites actively proliferate present > 90% viability on the third day.
- Harvesting amastigotes from BMDMs
- Remove the media from infected culture, add 5 ml of PBS and detach the cells using a cell scraper after one week of infection. Transfer the cell suspension to an ice cold glass Dounce tissue grinder and move the rod up and down 20 times to disrupt the cells, applying light pressure while maintaining the glass Dounce on ice. A large number of amastigotes are found in large parasitophorous vacuoles (Figure 1A), however the continuous parasite proliferation kills macrophages and gradually reduces parasite yield after purification.
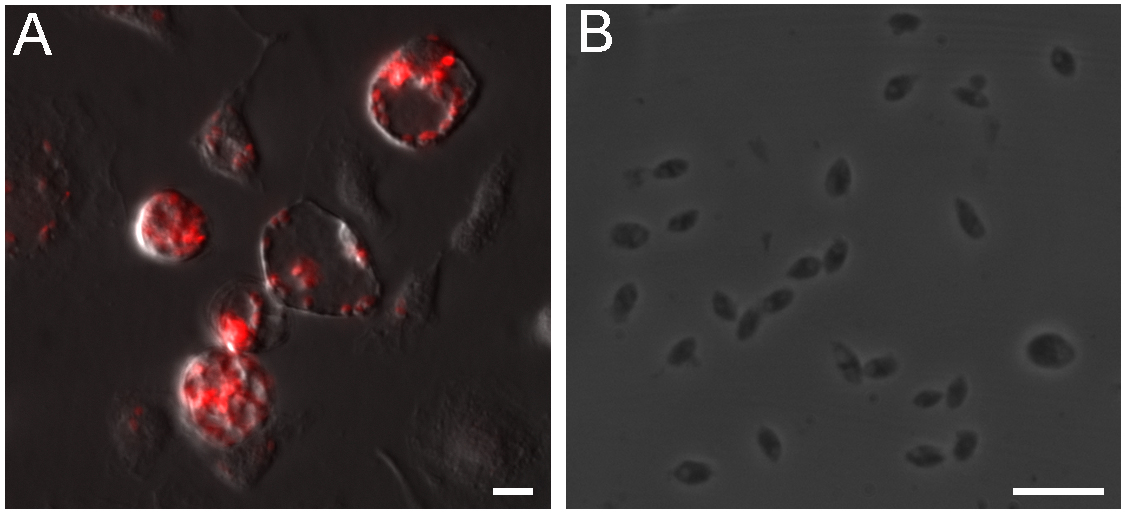
Figure 1. Intracellular L. amazonensis amastigotes isolation from macrophages. A. Balb/c BMDM infected with axenic amastigotes for 7 days, amastigotes expressing DsRed reside in large parasitophorous vacuoles. B. Clean amastigotes harvested from infected BMDMs. Scale bars = 10 μm. - Transfer the lysate to a 15 ml conical tube, complete the volume to 13 ml with cold PBS.
- Centrifuge at 210 x g for 8 min, 4 °C and transfer the supernatant to a fresh 15 ml conical tube. Discard the pellet, which contains intact macrophages and large debris.
- Centrifuge the harvested supernatant at 675 x g for 12 min, 4 °C.
- Discard the supernatant and resuspend the pellet in 3 ml of 25% Percoll cushion (750 μl of Percoll, 300 μl of FBS and 1.95 ml of RPMI 1640).
- Centrifuge at 3,000 x g for 15 min, 4 °C.
- Remove the supernatant with a serological pipette, resuspend the pellet in 3 ml of cold PBS and centrifuge at 800 x g for 12 min to wash the parasites.
- Resuspend the parasites in 1 ml of RPMI and count the amastigotes using a hemocytometer. 5-10 x 107 parasites should be obtained in a typical preparation. Figure 1B shows amastigotes obtained using this protocol.
- Transfer the suspension of parasites to a micro-centrifuge tube and centrifuge at 800 x g for 12 min.
- Resuspend the pellet in PBS to a concentration of 1.25 x 109 parasites/ml for fly injections.
- Remove the media from infected culture, add 5 ml of PBS and detach the cells using a cell scraper after one week of infection. Transfer the cell suspension to an ice cold glass Dounce tissue grinder and move the rod up and down 20 times to disrupt the cells, applying light pressure while maintaining the glass Dounce on ice. A large number of amastigotes are found in large parasitophorous vacuoles (Figure 1A), however the continuous parasite proliferation kills macrophages and gradually reduces parasite yield after purification.
- Fly injections
- Use 7 to 10 days old flies grown in wide fly vials (25 flies/vial) at 25 °C.
- Keep animals at 29 °C for 3 days before infection to optimize the UAS-GAL4 system efficiency.
- Prepare the needles using a capillary puller and measure the diameter of the tip using an electronic caliper, break the capillary needle to the diameter of approximately 60 μm. One quick tip is to make needles with approximately the diameter of the femur of the fly first pair of legs.
- Assemble the capillary needle to the Nanoject (Drumond) following manufacturer’s instructions.
Note: Use appropriate personal protective equipment and extreme caution to avoid accidents with the sharp glass needles. - Vortex the parasite suspension obtained in Procedure B for few seconds and load approximately 2 mm of the needle.
- Anesthetize the flies in a CO2 pad under a stereomicroscope.
- Adjust the injector to dispose of 32.2 nl in each injection. Load the microinjector with parasite suspension and inject the parasites in the lateral side of the fly, between the first and second abdominal segments (Video 1). Monitor the injection checking the expansion of the abdomen. Discard the flies that did not have noticeable inflation of their abdomen after 5 sec of injection.
- Transfer the injected insects to a fresh vial and close it tightly.
- Change the needle before starting a new vial to avoid cross contamination between flies of different vials.
- Prepare control groups injected with PBS and uninjected for each fly strain to unbiasedly evaluate the effect of infection on survival.Video 1. Injection of flies. This video demonstrates the injection of 32 nl of a 1% bromophenol blue solution (for better visualization) in adult flies using a microinjector. The dye solution enters the hemocoel and expands the abdomen.
- Use 7 to 10 days old flies grown in wide fly vials (25 flies/vial) at 25 °C.
- Quantification method 1: Survival assay
Wild-type flies are mildly susceptible to L. amazonensis infection, with 70-80% survival over the course of a 15-day experiment, but susceptible strains might present uncontrolled parasite proliferation in the hemolymph and reduced survival (Okuda et al., 2016).- Ideally, use at least 50 flies per condition, for statistical power. Distribute the 50 flies in 2 vials with cornmeal fly food. The yellow color of this meal allows easy detection of dead flies.
- Monitor for dead flies once daily for 10-15 days post-infection. Move surviving flies to new vials every 2-3 days or when many dead flies are sitting on the bottom of the vial, obscuring the counting process.
- Plot the data in Prism GraphPad using the survival function. The Kaplan-Meier survival curves of experimental flies should be compared to the same flies injected with PBS and to WT flies injected with the same preparation of parasites. The comparison can be performed using Log-rank (Mantel-Cox) test. Figure 2 shows the result of a typical survival experiment where flies lacking phagocytes also known as Phagoless (HmlΔGal4-eGFP, UAS-Bax, Defaye et al., 2009) are more susceptible than either WT flies challenged with parasites or Phagoless flies injected with PBS.
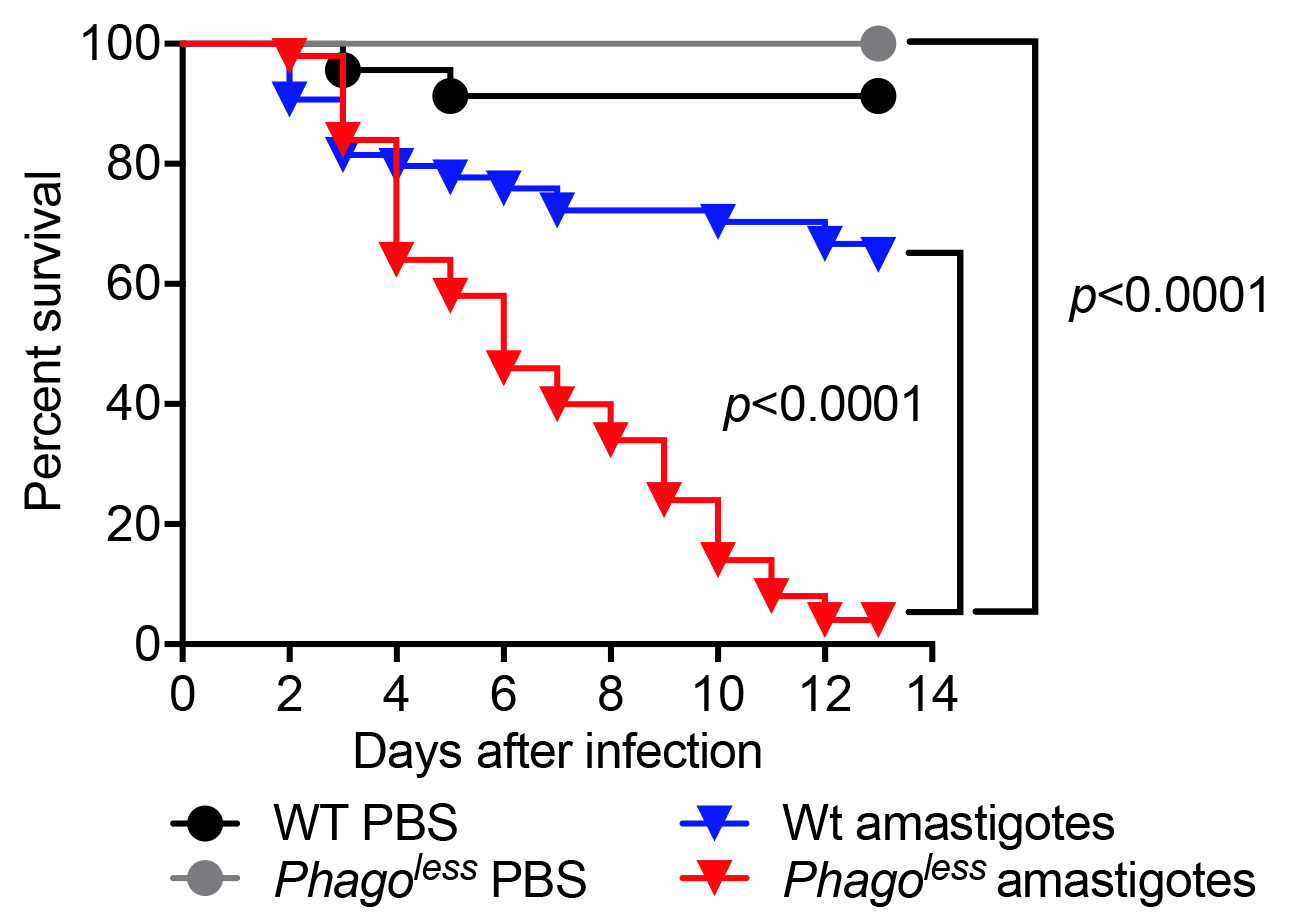
Figure 2. Survival of flies injected with L. amazonensis amastigotes. Flies devoid of hemocytes (Phagoless) present reduced resistance to Leishmania infection compared to wild-type flies. N = 60, statistical significance determined by Log-rank (Mantel-Cox test).
- Ideally, use at least 50 flies per condition, for statistical power. Distribute the 50 flies in 2 vials with cornmeal fly food. The yellow color of this meal allows easy detection of dead flies.
- Quantification method 2: Parasite burden determination by limiting dilution
In this assay, infected flies are homogenized and a fraction of the suspension is serially diluted in Schneider’s insect medium, cultivated for 7 days and the presence of proliferating promastigotes evaluated microscopically.- Fill three 96-well plates of with 100 μl of Schneider’s insect medium supplemented with 10% FBS, 50 U/ml of a penicillin/streptomycin solution, 10 µg/ml tetracycline and 5 µg/ml of tunicamycin (DsRed parasites are tunicamycin resistant). Add an additional 80 µl of media to the first row of one plate that will be the first dilution well. The high dose of antibiotics is essential to impair the growth of the bacteria present in the flies.
- Inside the hood, using a fine tweezer, pick one fly from the CO2 pad and quickly dip it into 70% ethanol, and then dip twice in sterile water. Put the clean fly in a 1.5 ml micro-centrifuge tube containing 200 µl of PBS.
- Gently grind the fly for 10 sec using a pellet grinder to disrupt the exoskeleton and release parasites from the hemolymph and tissues.
- Wait 5 min to settle the large tissues in the bottom and transfer 20 µl of the top fly homogenate into the first well in triplicate.
- Using a multichannel pipette make the serial dilution by transferring 100 µl of one well to the next well down the column, for 24 dilutions (until all the wells of the 3 plates are utilized). Change the pipette tips in each of the first 5 dilutions and every 3 dilutions afterwards to avoid carryover of parasites in the tips.
- After 7 days at 27 °C, check for the presence of live promastigotes under a light microscopy. Count the number of positive wells in each series and convert the value to the dilution factor (first well = 1/20 of one fly).
Tip: During incubation, keep one side of the plate about 5 mm higher so the parasites will concentrate in the same side of the well to facilitate the identification of positive wells.
- Fill three 96-well plates of with 100 μl of Schneider’s insect medium supplemented with 10% FBS, 50 U/ml of a penicillin/streptomycin solution, 10 µg/ml tetracycline and 5 µg/ml of tunicamycin (DsRed parasites are tunicamycin resistant). Add an additional 80 µl of media to the first row of one plate that will be the first dilution well. The high dose of antibiotics is essential to impair the growth of the bacteria present in the flies.
- Quantification method 3: Direct visualization of parasites from infected flies
This method can be used to estimate the parasite burden of flies infected with parasites expressing fluorescent proteins such as DsRed.- Gently grind a single fly in 40 µl of Schneider’s insect medium in a 1.5 ml micro-centrifuge tube using a pellet grinder for 10 sec and let it stand for 5 min to precipitate large debris. We suggest using at least 10 flies per sample for statistical power.
- Load 2 improved Neubauer or similar disposable counting chambers with the top suspension.
- Image the DsRed-expressing parasites using a fluorescence microscope with a 10x objective. Take 4 images per chamber for parasite counting (8 images in total).
- Using a cell image analysis software such as the open source CellProfiler (Carpenter et al., 2006) adjust the settings to precisely identify the parasites according to the size and fluorescence. An example of a simple pipeline for Cell Profiler is shown in Figure 3.
- Multiply the average number of parasites per image by 1.82 x 104 to calculate the number of parasites per ml, then multiply by 0.04 (40 μl total volume) to calculate the total number of parasites per fly. Adjust this formula if using a different imaging setup, considering the actual size of the image and the depth of the counting chamber.
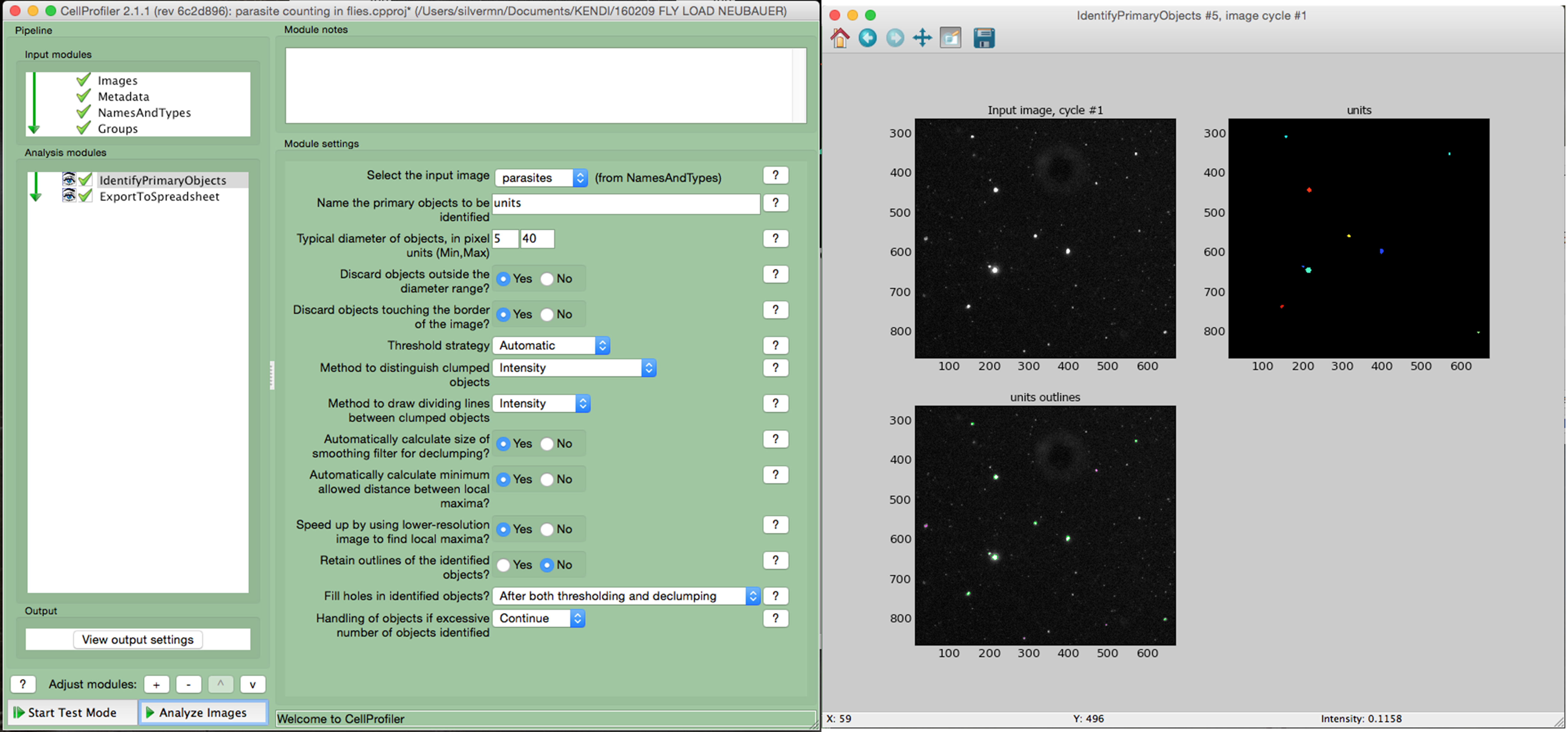
Figure 3. Counting parasites in fly homogenates using CellProfiler. The homogenate of a fly infected for 2 days was loaded into a counting chamber and imaged using a fluorescence microscope. The images were analyzed using Cell Profiler software. The left window shows the settings used to identify the parasites in the image. The right window shows the input raw image, the image with the identified parasites (colored) and, at the bottom, the image highlights all the identified objects (parasites are circled in green).
- Gently grind a single fly in 40 µl of Schneider’s insect medium in a 1.5 ml micro-centrifuge tube using a pellet grinder for 10 sec and let it stand for 5 min to precipitate large debris. We suggest using at least 10 flies per sample for statistical power.
Data analysis
- For fly survival experiments, experimental flies are compared to wild-type flies with the same genetic background injected with same parasite preparation and to flies injected with PBS (vehicle). This detail is important because some mutant flies can be susceptible to injury and die after the injection of PBS. The statistical significance is determined by Log-rank (Mantel-Cox test).
- For parasite load determination by limiting dilution and by direct counting, the replicates of each 10 experimental flies are compared to WT flies infected with the same parasite preparation using One-way ANOVA.
- We perform at least 3 independent experiments for in vivo assays. Additionally, fly lines presenting phenotype of interest are validated by 3 additional independent assays.
- Statistical test was performed with GraphPad Prism7. Freeware such as R can also be used to perform this type of analysis.
Recipes
- 199 media to grow promastigotes
199 media supplemented with:
10% FBS
20 mM HEPES
0.35 g/L sodium bicarbonate
5 μg/ml hemin (from a stock solution of 0.2% hemin in 0.1 N NaOH), pH 7.4 - 199 media to induce promastigote to axenic amastigote differentiation
199 media supplemented with:
20% FBS
20 mM HEPES
0.35 g/L sodium bicarbonate
5 μg/ml hemin (from a stock solution of 0.2% hemin in 0.1 N NaOH)
40 mM sodium succinate
0.5% tryptic soy broth, pH 5.4
Acknowledgments
This protocol has been adapted from Okuda et al., 2016. Supported by the NIH (R21AI109678) to NS. The authors declare that there is no conflict of interest.
References
- Brand, A. H. and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118(2): 401-415.
- Carpenter, A. E., Jones, T. R., Lamprecht, M. R., Clarke, C., Kang, I. H., Friman, O., Guertin, D. A., Chang, J. H., Lindquist, R. A., Moffat, J., Golland, P. and Sabatini, D. M. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7(10): R100.
- Chen, R. (2013). Isolation and culture of mouse bone marrow-derived macrophages (BMM'phi'). Bio Protoc e68.
- Defaye, A., Evans, I., Crozatier, M., Wood, W., Lemaitre, B. and Leulier, F. (2009). Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J Innate Immun 1(4): 322-334.
- Dutta, S., Ray, D., Kolli, B. K. and Chang, K. P. (2005). Photodynamic sensitization of Leishmania amazonensis in both extracellular and intracellular stages with aluminum phthalocyanine chloride for photolysis in vitro. Antimicrob Agents Chemother 49(11): 4474-4484.
- Gupta, N., Goyal N. and Rastogi, A. K. (2001). In vitro cultivation and characterization of axenic amastigotes of Leishmania. Trends Parasitol 17(3) 150-153.
- Okuda, K., Tong, M., Dempsey, B., Moore K.J., Gazzinelli, R.T. Silverman, N. (2016). Leishmania amazonensis engages CD36 to drive parasitophorous vacuole maturation. PLoS Pathog 12(6): e1005669.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Okuda, K. and Silverman, N. (2017). Drosophila Model of Leishmania amazonensis Infection. Bio-protocol 7(23): e2640. DOI: 10.21769/BioProtoc.2640.
Category
Immunology > Animal model > Drosophila (Fruit fly)
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











