- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Bacterial Polyhydroxybutyrate Content by Flow Cytometry
Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2638 Views: 8133
Reviewed by: Valentine V TrotterPaula María TribelliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1764 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1808 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1779 Views
Abstract
We describe here a detailed protocol for the quantification of the intracellular content of polyhydroxybutyrate (PHB) in a population of bacterial cells by flow cytometry, which is based on a consensus of several previously reported works.
Keywords: PolyhydroxybutyrateBackground
Upon nutrient exhaustion and in the presence of an available carbon source, several bacterial species (e.g., Cupriavidus necator, Sinorhizobium meliloti, species of Pseudomonas) switch on the accumulation of the neutral polymer of β-hydroxybutyrate, polyhydroxybutyrate (PHB), for storage of carbon and reducing power (Jendrossek and Pfeiffer, 2014). Classical methods for quantification of bacterial PHB content are based on its initial depolymerization, followed by the quantitative detection of specific chemical derivatives of β-hydroxybutyrate by UV spectrophotometry or gas chromatography (Law and Slepecky, 1961; Braunegg et al., 1978). More recently, the application of the fluorochrome Nile red (9-diethylamino-5H-benzo[α]phenoxazine-5-one) (Greenspan and Fowler, 1985) to stain intracellular PHB granules (Müller et al., 1993) led to the development of novel rapid fluorometric methods to quantify the content of the polymer in bacterial samples. Subsequently, coupling of Nile red–based PHB staining to flow cytometry made possible the quantification of the polymer content at a single-cell level within bacterial populations (Gorenflo et al., 1999; Kacmar et al., 2006; Tyo et al., 2006; Alves et al., 2017; Lagares Jr. et al., 2017). We report here a detailed protocol that summarizes the consensus steps for the quantification of PHB in bacterial cells by Nile red staining coupled to flow cytometry.
Materials and Reagents
- Pipette tips (2-200 µl and 100-1,000 µl tips)
- 1.5 ml Eppendorf tubes
- 12 x 75-mm Falcon® 5 ml uncapped Round Bottom Polystyrene Test tubes (Corning, catalog number: 352052 )
- Bacterial samples for analysis
Note: The bacteria can be harvested from multiple sources (e.g., liquid cultures, colonies on semisolid agar plates, natural environments). However, it must be taken into account that adaptations in the steps for cell preparation should be considered, depending on the specific source of the cells. - Absolute ethanol (e.g., Sigma-Aldrich, catalog number: E7023 )
- Sterile deionized water (e.g., Sigma-Aldrich, catalog number: W3500 )
- Sodium chloride (NaCl) (e.g., Sigma-Aldrich, catalog number: S9888 )
- Potassium chloride (KCl) (e.g., Sigma-Aldrich, catalog number: P3911 )
- Sodium phosphate dibasic (Na2HPO4) (e.g., Sigma-Aldrich, catalog number: S9763 )
- Potassium phosphate monobasic (KH2PO4) (e.g., Sigma-Aldrich, catalog number: P0662 )
- Dimethyl sulfoxide (DMSO) (e.g., Sigma-Aldrich, catalog number: D8418 )
- Nile red stain (e.g., Sigma-Aldrich, catalog number: N3013 )
- 1 N HCl and 1 N NaOH solutions for pH adjustments
- Cytometer sheath fluid (e.g., BD FACSFlowTM solution, BD, catalog number: 342003 )
- Full-strength bleach (for waste disposal)
- Phosphate-buffered saline (PBS; see Recipes for 10x and 1x solution preparation)
- Permeabilization solution (see Recipes)
- Nile red stock solution (see Recipes)
Equipment
- Pipets (e.g., Gilson Pipetman Classic) (Gilson, models: P20 , P200 , P1000 )
- Centrifuge for 1.5 ml Eppendorf tubes (e.g., Eppendorf, model: 5424 , catalog number: 5424000010)
- pH meter
- Spectrophotometer suitable for measurement of optical density at 600 nm (OD600)
- FACSCalibur flow cytometer (BD, model: FACSCALIBUR ) equipped with an excitation laser of 488-nm-wavelength and an emission filter with bandwidth of 585 ± 45 nm
Note: The suitability of other flow cytometers than FACSCalibur (BD, USA) for this assay is subjected to the availability of a proper excitation laser source and of the filter for the emitted fluorescence, as indicated above.
Software
- Flow cytometer acquisition software
- FlowJo [FlowJo, USA]
Procedure
- Bacterial PHB staining
- Pellet down ca. 5 x 108 bacterial cells by centrifugation at 7,500 x g for 5 min at room temperature (ca. equivalent to 1 ml of an Escherichia coli liquid culture at an OD600 of 0.5).
Notes:- Note that lower amounts of cells will be subjected to the flow cytometric analysis, however, higher initial bacterial amounts are easier to handle during the centrifugation steps prior to staining.
- Include as a negative control for PHB measurements a bacterial population (ideally of the same species) grown under conditions that are not permissive for PHB accumulation (here referred to as PHB- bacteria), and a population of bacteria loaded with PHB (PHB+ bacteria) that has not been stained with Nile red, as a negative control for non-specific fluorescence.
- Centrifugation speed and time need to be adjusted in each particular case to optimize bacterial collection at the bottom of the tube, mainly depending on the viscosity of the supernatant.
- Note that lower amounts of cells will be subjected to the flow cytometric analysis, however, higher initial bacterial amounts are easier to handle during the centrifugation steps prior to staining.
- Wash the bacterial pellet once with 500 µl 1x PBS (see Recipes), and resuspend cells in 500 µl of permeabilization solution (see Recipes). Incubate the suspension at room temperature (RT) for 15 min.
- Collect the cells by centrifugation at 7,500 x g for 5 min at RT.
- Resuspend the pellet in 1 ml 1x PBS and dilute the bacterial suspension 1:100 with 1x PBS to a final volume of 1 ml (i.e., to reach a bacterial concentration of ca. 5 x 106 cells per milliliter). Mix well by vortexing for a few seconds.
- Add 20 µl of Nile red stock solution (see Recipes) to the diluted bacterial suspension. Mix well by vortexing for a few seconds and incubate protected from light at RT for 30 min.
Note: Please note that, as reviewed by Alves et al. (2017), the concentration of Nile red that is required to optimally stain the bacterial PHB granules largely varies among species (i.e., from 0.032 µM for C. necator to 94.2 µM for E. coli). The concentration reported in this protocol has been shown to properly stain PHB in Sinorhizobium meliloti (Lagares Jr. et al., 2017). Nile red concentration and exposure time should be optimized for each bacterial species. - Transfer samples to 12 x 75-mm 5 ml Falcon® test tubes and proceed to analyze them by flow cytometry as indicated in the following steps.
Note: Since Nile red fluorescence starts to decay shortly after its addition (Alves et al., 2017), it is critical to measure all the samples at an equal time after they are stained.
- Pellet down ca. 5 x 108 bacterial cells by centrifugation at 7,500 x g for 5 min at room temperature (ca. equivalent to 1 ml of an Escherichia coli liquid culture at an OD600 of 0.5).
- Flow cytometry
- Set the flow cytometer for cell excitation by the 488-nm-wavelength argon laser and Nile red fluorescence acquisition by the filter with bandwidth centered at 585 nm (on FL2 channel if using FACScalibur flow cytometer). Register Forward (FSC)- and side (SSC)-scattered light as well. Adjust FSC, SSC and FL2 detector voltages, if necessary. Set the sample flow rate at the low flow position (e.g., ca. 12 µl/sec).
Notes:- Carefully follow flow cytometer facility use guidelines (e.g., warm-up instructions, sheath and waste management).
- In general, setting FSC and SSC channel voltages to E02 and 300, respectively, would allow to clearly identify bacterial populations in FSCxSSC dot plots. FL2 channel voltage should be adjusted to maximize the difference of the median values corresponding to PHB+ and PHB- bacterial populations (e.g., FL2 channel voltage was set to 500 during acquisition of the data plotted in Figure 1).
- Carefully follow flow cytometer facility use guidelines (e.g., warm-up instructions, sheath and waste management).
- Analyze samples and controls with the flow cytometer. Record at least 100,000 events per sample.
- Set the flow cytometer for cell excitation by the 488-nm-wavelength argon laser and Nile red fluorescence acquisition by the filter with bandwidth centered at 585 nm (on FL2 channel if using FACScalibur flow cytometer). Register Forward (FSC)- and side (SSC)-scattered light as well. Adjust FSC, SSC and FL2 detector voltages, if necessary. Set the sample flow rate at the low flow position (e.g., ca. 12 µl/sec).
Data analysis
Analyze data with a suitable software (e.g., FlowJo [FlowJo, USA]). The median fluorescence intensity (MFI; expressed in AFU, Arbitrary Fluorescence Units) from histograms of FL2-H has been reported to linearly correlate well with the bacterial PHB content (Gorenflo et al., 1999; Kacmar et al., 2006; Tyo et al., 2006; Ratcliff et al., 2008; Alves et al., 2017). The population of bacteria loaded with PHB in each sample could be identified by comparing the corresponding MFI with that of the bacteria that were analyzed as a negative control (see Figure 1). In bacterial samples displaying phenotypic heterogeneity, the percentage of PHB+ cells could be calculated. At least three independent samples must be processed as biological replicates.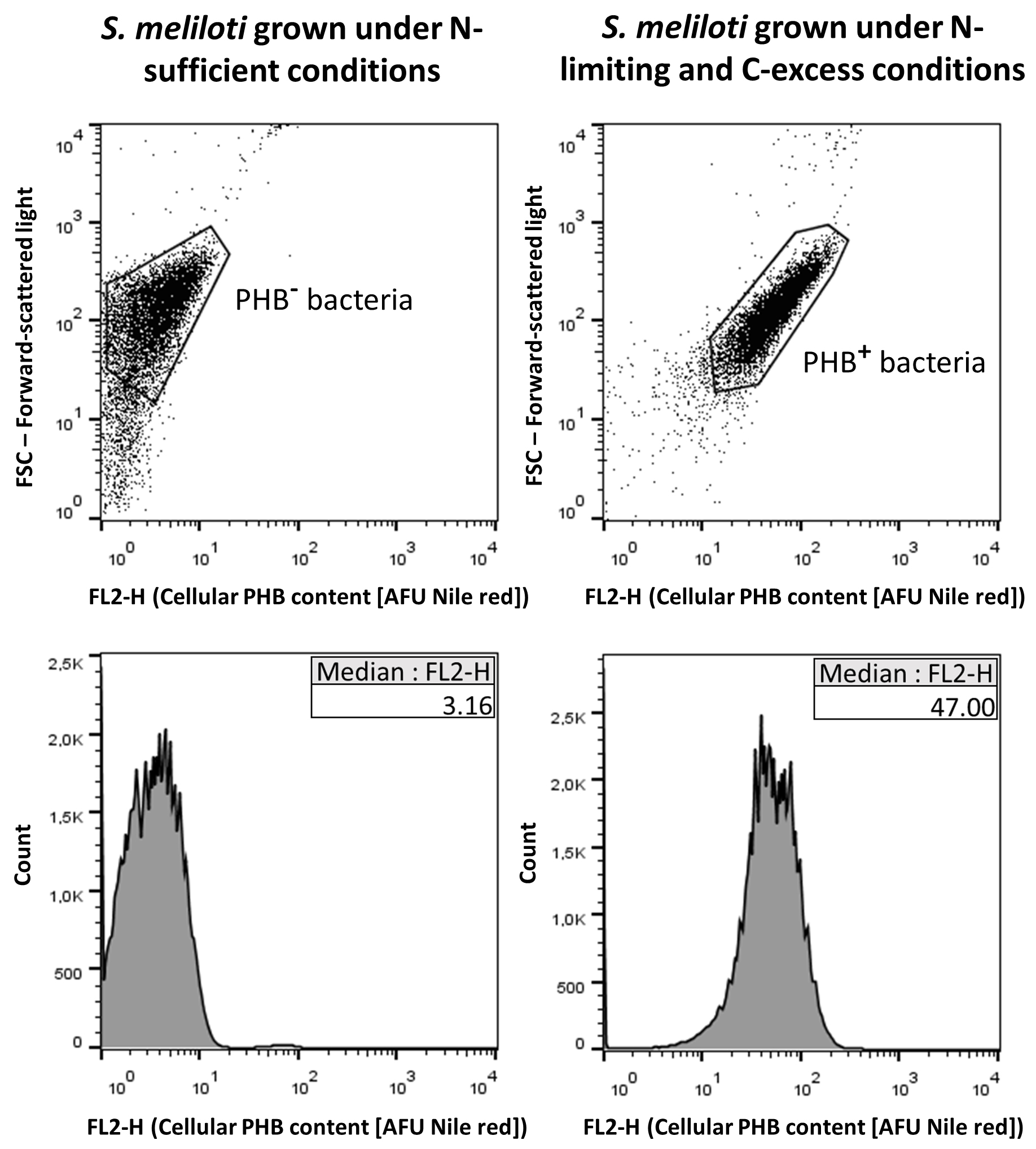
Figure 1. Example of dot plots (upper panels) and histograms (lower panels) from a sample of Sinorhizobium meliloti cells grown under a nitrogen-limiting and carbon-excess condition that enables PHB accumulation (right panels, PHB+ bacteria), and from a sample of the same bacterial species grown under nitrogen sufficient conditions as a negative control for PHB accumulation (left panels, PHB- bacteria) (Lagares Jr. et al., 2017). The x-axis represents the cellular PHB content estimated by the fluorescence of Nile red (expressed in AFU, Arbitrary Fluorescence Units) that was registered by the channel FL2. The median fluorescence intensity (MFI) values for each bacterial population are indicated in the corresponding histogram insets.
Recipes
- Phosphate-buffered saline (PBS) (10x concentrated)
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4
0.24 g KH2PO4
Adjust pH to 7.2 with 1 N NaOH or 1 N HCl and make up to 100 ml with water (usually less than 1 ml of 1 N NaOH or 1 N HCl stocks would be enough)
Autoclave and store at RT - Permeabilization solution (ethanol 35% in PBS [vol/vol])
350 µl absolute ethanol
100 µl 10x PBS
550 µl sterile deionized water - Nile red stock solution
1 mg Nile red
Make up to 1 ml with DMSO
Store at 4 °C
Acknowledgments
This work was supported by the National Scientific and Technical Research Council (CONICET), the National Agency for Promotion of Science and Technology (ANPCyT), and Universidad Nacional de Quilmes. A.L. was supported by CONICET fellowships. C.V. is a researcher of CONICET. The current protocol was adapted from (Lagares Jr. et al., 2017), which was mainly based on several previously reported works (Kacmar et al., 2006; Ratcliff et al., 2008). The authors declare no conflicts of interest.
References
- Alves, L. P., Almeida, A. T., Cruz, L. M., Pedrosa, F. O., de Souza, E. M., Chubatsu, L. S., Muller-Santos, M. and Valdameri, G. (2017). A simple and efficient method for poly-3-hydroxybutyrate quantification in diazotrophic bacteria within 5 minutes using flow cytometry. Braz J Med Biol Res 50(1): e5492.
- Braunegg, G., Sonnleitner, B. and Lafferty, R. M. (1978). A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Appl Microbiol Biotechnol 6: 29-37.
- Gorenflo, V., Steinbuchel, A., Marose, S., Rieseberg, M. and Scheper, T. (1999). Quantification of bacterial polyhydroxyalkanoic acids by Nile red staining. Appl Microbiol Biotechnol 51(6): 765-772.
- Greenspan, P. and Fowler, S. D. (1985). Spectrofluorometric studies of the lipid probe, nile red. J Lipid Res 26(7): 781-789.
- Jendrossek, D. and Pfeiffer, D. (2014). New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16(8): 2357-2373.
- Kacmar, J., Carlson, R., Balogh, S. J. and Srienc, F. (2006). Staining and quantification of poly-3-hydroxybutyrate in Saccharomyces cerevisiae and Cupriavidus necator cell populations using automated flow cytometry. Cytometry A 69(1): 27-35.
- Lagares, A., Jr., Ceizel Borella, G., Linne, U., Becker, A. and Valverde, C. (2017). Regulation of polyhydroxybutyrate accumulation in Sinorhizobium meliloti by the trans-encoded small RNA MmgR. J Bacteriol 199(8).
- Law, J. H. and Slepecky, R. A. (1961). Assay of poly-β-hydroxybutyric acid. J Bacteriol 82: 33-36.
- Müller, S., Lösche, A. and Bley, T. (1993). Staining procedures for flow cytometric monitoring of bacterial populations. Acta Biotechnol 13: 289-297.
- Ratcliff, W. C., Kadam, S. V. and Denison, R. F. (2008). Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiol Ecol 65(3): 391-399.
- Tyo, K. E., Zhou, H. and Stephanopoulos, G. N. (2006). High-throughput screen for poly-3-hydroxybutyrate in Escherichia coli and Synechocystis sp. strain PCC6803. Appl Environ Microbiol 72(5): 3412-3417.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lagares Jr., A. and Valverde, C. (2017). Quantification of Bacterial Polyhydroxybutyrate Content by Flow Cytometry. Bio-protocol 7(23): e2638. DOI: 10.21769/BioProtoc.2638.
Category
Microbiology > Microbial metabolism > Other compound
Microbiology > Microbial biochemistry > Other compound
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









