- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
zPACT: Tissue Clearing and Immunohistochemistry on Juvenile Zebrafish Brain
Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2636 Views: 12616
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Various Modes of Spinal Cord Injury to Study Regeneration in Adult Zebrafish
Subhra Prakash Hui and Sukla Ghosh
Dec 5, 2016 13377 Views

Detection of mRNA by Whole Mount in situ Hybridization and DNA Extraction for Genotyping of Zebrafish Embryos
Rachna Narayanan and Andrew C. Oates
Mar 20, 2019 15724 Views

Qualitative Detection of Lipid Peroxidation in Mosquito Larvae Using Schiff’s Reaction: A Simple Histochemical Tool for In Situ Assessment of Oxidative Damage
Antonella Cuniolo [...] María Victoria Martin
Feb 5, 2026 54 Views
Abstract
In studies of brain function, it is essential to understand the underlying neuro-architecture. Very young zebrafish larvae are widely used for neuroarchitecture studies, due to their size and natural transparency. However, this model system has several limitations, due to the immaturity, high rates of development and limited behavioral repertoire of the animals used.
We describe here a modified version of the passive clearing technique (PACT) (Chung et al., 2013; Tomer et al., 2014; Yang et al., 2014; Treweek et al., 2015), which facilitates neuroanatomical studies on large specimens of aquatic species. This method was initially developed for zebrafish (Danio rerio) (Frétaud et al., 2017; Mayrhofer et al., 2017; Xavier et al., 2017), but has also been successfully tested on other fish, such as medaka (Oryzias latipes) (Dambroise et al., 2017), Mexican cave fish (Astyanax mexicaus) and African zebra mbuna (Metriaclima zebra), and on other aquatic species, such as Xenopus spp. (Xenopus laevis, Xenopus tropicalis) (Fini et al., 2017). This protocol, based on the CLARITY method developed and modified by Deisseroth’s laboratory and others (Chung et al., 2013; Tomer et al., 2014; Yang et al., 2014), was adapted for use in aquatic species, including zebrafish in particular (zPACT).
This protocol is designed to render zebrafish specimens optically transparent while preserving the overall architecture of the tissue, through crosslinking in a polyacrylamide/formaldehyde mesh. Most of the lipids present in the specimen are then removed by SDS treatment, to homogenize the refractive index of the specimen by eliminating light scattering at the water/lipid interface, which causes opacity. The final clearing step, consists of the incubation of the specimen in a fructose-based mounting medium (derived from SeeDB) (Ke et al., 2013), with a refractive index matching that of the objective lens of the microscope. The combination of this technique with the use of genetically modified zebrafish in which green fluorescent protein (GFP) is expressed in specific cell populations provides opportunities to describe anatomical details not visible with other techniques.
Background
Adult teleost fish (zebrafish, medaka) are increasingly being used as model vertebrates in studies extending from stem cells to brain development (Than-Trong and Bally-Cuif, 2015; Dambroise et al., 2017), in all aspects of physiological and medical research (Oksanen et al., 2013) and in studies of complex types of behavior, such as learning, memory and addiction (Bailey et al., 2015).
In its role as a scientific service provider, the Tefor Core Facility has developed a reliable tissue clearing technique for dense, neuronal tissues of several millimeters in diameter, to satisfy the demand for studies of large specimens of aquatic animals.
By creating a set of neuroanatomical atlases of different developmental stages, this protocol will serve as a resource for other laboratories wishing to integrate their data into these anatomical reference frameworks (ARFs). At the time of publication, the prototype of a 7dpf ARF is available from https://www.zebrafish.tefor.net/. The data accessible via this URL will subsequently be consolidated into an annotated 3D neuroanatomy atlas and enriched with standardized high-resolution confocal imaging data for different developmental stages. The Tefor Core Facility is currently developing ARFs for 5 dpf, 6 dpf, 7 dpf larval heads and juvenile brains. The development of ARFs for other stages will depend on the demand from members of the zebrafish research community.
The integration of new specimens into the corresponding ARF requires precise staging of the developmental stage of the specimen and the use of DiI stain, as described here. This dye is used as a fiducial marker for the computational alignment of 3D imaging data into the corresponding ARF. However, the protocol can also be run without these steps if a direct comparison of the generated data with the existing data pool is not required.
As a compromise between specimen size and maturity, we focused on juvenile zebrafish brains during the development of this protocol. Juvenile zebrafish brains are smaller than those from older fish, but may be considered mature (Filippi, 2010). Moreover, the use of sexually immature animals should reduce data variability due to sexual dimorphism (Ampatzis, 2012). The smaller size of these animals also makes the protocol faster, because all its steps are dependent on the penetration of the applied agents into the specimens. However, the animals used have nevertheless reached a sufficiently advanced developmental stage for extrapolation of the findings to older specimens.
Materials and Reagents
- Sterile 50 ml conical tubes (Corning, catalog number: 430291 )
- Sharp blade (single-sided razor blade (e.g., Razor Blade Company, catalog number: 62-0167 ) or appropriate scalpel blade (e.g., #10))
- 94 x 16 mm Petri dishes (Greiner Bio One International, catalog number: 632102 )
- Small spatula for manipulating anesthetized fish during staging
- Sterile 15 ml conical tubes (Corning, catalog number: 431470 )
- Paintbrush (e.g., Kolinsky France, Manet 630, size 3 )
- 60 x 15 mm Petri dishes (Greiner Bio One International, catalog number: 628160 )
- Multipolymer protective gloves (Shield Scientific, catalog number: 66 9253 )
- Parafilm M (Sigma-Aldrich, Parafilm, catalog number: P7543 )
- Glass tubes for a rotisserie hybridization oven (Fisher Scientific, catalog number: 11791436 )
- 2 ml Eppendorf tube (STARLAB INTERNATIONAL, catalog number: E1420-2000 )
- 3.2 ml transfer pipette (Carl Roth, catalog number: EA67.1 )
- 2 ml glass tubes (Carl Roth, catalog number: H303.1 )
- 0.22 µm-pore filter (SARSTEDT, catalog number: 83.1826.001 )
- 6-week-old zebrafish raised under standard conditions (density, feeding, etc.; Lawrence and Mason, 2012; Lawrence et al., 2016)
- Rotifers (Brachionus plicatilis)
- Brine shrimps (Artemia nauplii)
- Dry food (Skretting, Gemma Micro)
- Pure nitrogen (Air Liquide, ALPHAGAZ 1)
- Alexa Fluor 488-conjugated secondary antibody, goat anti-chicken IgY (H+L) (Thermo Fisher Scientific, InvitrogenTM, catalog number: A-11039 )
- DiIC18(3) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D282 )
- Dimethyl sulphoxide (DMSO) (Carl Roth, catalog number: 4720.1 )
- Primary antibody directed against GFP (chicken) (Aves Lab, catalog number: GFP-1020 )
- Low-melting point agarose (Sigma-Aldrich, catalog number: A4018 )
- Mineral oil (Carl Roth, catalog number: HP50.3 )
- Tricaine mesylate (MS222) (Sigma-Aldrich, catalog number: A5040 )
- Sodium bicarbonate (Carl Roth, catalog number: 0965.2 )
- 20x PBS (Santa Cruz Biotechnology, catalog number: sc-362183 )
- 16% formaldehyde (w/v), methanol-free (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 28908 )
- 4% formaldehyde solution, methanol-free (w/v) (PFA) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 28908 )
- Tween 20 (Sigma-Aldrich, catalog number: P7949 )
- 40% acrylamide solution in H2O (Sigma-Aldrich, catalog number: 01697 )
- VA-044 initiator (Wako Pure Chemical Industries, catalog number: 017-19362 )
- SDS pellet (Carl Roth, catalog number: CN30.3 )
- Boric acid (Carl Roth, catalog number: 6943.1 )
- 20x SSC (Carl Roth, catalog number: 1054.1 )
- Formamide (CH3NO) (Carl Roth, catalog number: 6749.3 )
- H2O2 solution (30%) (Carl Roth, catalog number: CP26.5 )
- Sodium azide (Sigma-Aldrich, catalog number: S2002 )
- Normal goat serum (NGS) (Antibodies, catalog number: 54-410 )
- Triton X-100 (Fisher Scientific, catalog number: BP151-500 )
- D(-)-fructose (Carl Roth, catalog number: 4981.6 )
- SYLGARD 184 (Sigma-Aldrich, catalog number: 761036 )
- 10x MS222 solution (see Recipe 1)
- Fixing solution (4% PFA, 0.01 M PBS, 0.1% Tween 20) (see Recipe 2)
- Sylgard-coated Petri dish (see Recipe 3)
- PBST (1x PBS, 0.01% Tween 20) (see Recipe 4)
- Hydrogel solution (see Recipe 5)
- SDS solution (8% SDS, 0.2 M boric acid, pH 8.5) (see Recipe 6)
- Depigmentation (see Recipe 7)
- SSC 0.5x-Tween 20
- Depigmentation solution
- SSC 0.5x-Tween 20
- Storage solution (see Recipe 8)
- Blocking solution (see Recipe 9)
- Staining solution (see Recipe 10)
- Fructose-based high-refractive index solution (fbHRI) (see Recipe 11)
Equipment
- 3 fish tanks/line (e.g., Tecniplast 1 L Breeding Tank without the internal part)
- Laminated measuring grid (see Figure 1)
- Fume hood
- Ice bath & refrigerator
- Embryo spoon (Carl Roth, catalog number: TL85.1 )
- Dumont tweezers, straight (World Precision Instruments, catalog number: 500336 )
- Dumont tweezers, straight (World Precision Instruments, catalog number: 14099 )
- Dumont tweezers, 45° angle (World Precision Instruments, catalog number: 500234 )
- Stereomicroscope (Olympus, model: SZX10 )
- Fiber light source (Olympus, model: KL 2500 LED )
- Heated vacuum desiccator (VWR, SELECTA, catalog number: SELE4000474 )
- Hotplate (Labexchange, Bioblock Scientific, model: AM3001K )
- Vacuum pump (Savant, model: Gel Pump GP 110 )
- Histology processing cassettes (Simport, model: M503-12 )
- 400 ml beaker (VWR, catalog number: 213-1125 )
- 600 ml beaker (VWR, catalog number: 213-1126 )
- Rotisserie hybridization oven (Grant Boekel, model: HIR 1011 )
- 3D rocker Polymax 1040 (Heidolph Instruments, catalog number: 0 36130320 )
- Refractometer (KRUSS Optronic, catalog number: DR 301-95 )
- Leica TCS SP8 laser scanning microscope (Leica Microsystems, model: Leica TCS SP8 )
- 2 photomultipliers for the simultaneous detection of two channels
- Leica HC FLUOTAR L 25x/1.00 IMM motCorr objective (Leica Microsystems, catalog number: 15507703 )
- 2 photomultipliers for the simultaneous detection of two channels
- Heavy magnetic stirring bar (SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F37118-0002 )
Software
- Fiji/ImageJ (http://fiji.sc)
- Amira (https://www.fei.com/software/amira-3d-for-life-sciences/)
- ITK-snap (http://www.itksnap.org)
Procedure
- Rearing and staging
Fish are reared for 6 weeks under controlled conditions (photoperiod = 14 h/10 h light/dark; temperature = 26.5 ± 1 °C; pH = 7.8 ± 0.1; conductivity = 240 ± 30 µS/cm, NH4+ = 0 mg/L, NO2- = 0 mg/L, NO3- < 50 mg/L), in groups of 50 individuals per 1.4 L tank. They are fed on rotifers (Brachionus plicatilis; > 1,000/fish/day; one dose at noon) for the first two weeks of feeding, and then with brine shrimps (Artemia nauplii; ~250/fish/day) and dry food (Skretting, Gemma Micro; to apparent satiation, two doses, morning, afternoon) (Lawrence and Mason, 2012; Lawrence et al., 2016). All procedures are performed in accordance with European Union Directive 2010/63/EU.
As suggested in a previous study (Parichy et al., 2009), the stage of interest (juvenile) is defined on the basis of the disappearance of the fin-fold, at about 5-6 weeks in local standard conditions, corresponding to a body length of 12.5 mm.
The measuring and staging steps are performed under potentially lethal anesthesia and outside of water. These steps must therefore be performed rapidly, individually and very carefully, to ensure that all the fish survive the procedure. Fish of the desired size should be killed directly before fixation, to prevent a decay of expression patterns in the specimens. Measuring/staging should be performed in the fish facility, so these animals must be kept alive until they are transferred to the lab and killed and fixed under a fume hood. Fish that are too small can be left for a few days and then remeasured, and fish that are too large can be introduced into other experiments or the breeding program.- Prepare the anesthetic solution: 200 ml of 1x MS222 solution, by diluting 10x MS222 solution (see Recipe 1) 1:9 in fish water in a fish tank (anesthesia tank).
- For each fish line, prepare two additional fish tanks:
- Name of the fish line + correct size.
- Name of the fish line + incorrect size.
- Name of the fish line + correct size.
- Anesthetize one fish at a time by transferring it to the anesthesia tank.
Wait until the fish is completely anesthetized; test this by touching the tail with a spatula and checking for movement. - Position the fish directly on the measuring grid (Figure 1).
- Remove the liquid surrounding the fish to avoid lens effects.
- With the spatula, position the mouth of the fish against the gray middle line and the back along the horizontal line, as shown in Figure 1.
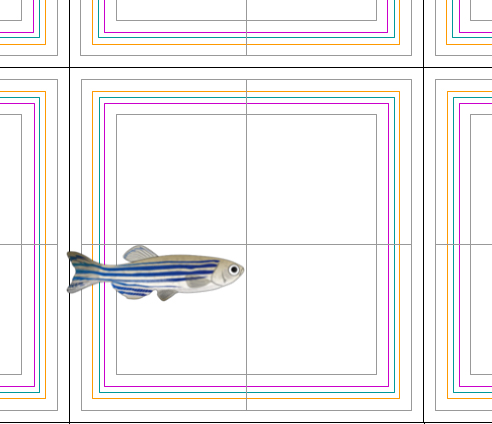
Figure 1. Staging juvenile fish by age and measuring their body length: Diagram of a fish on the measuring grid. The different color lines correspond to different distances from the center lines of the measuring grid: inner gray line 11 mm; magenta line 12 mm; cyan line 12.5 mm; yellow line 13 mm; outer gray line 14 mm. The fish in this figure would be considered to have a body length of 12.5 mm (central gray line to the cyan line). Zebrafish image modified from Togo picture gallery maintained by Database Center for Life Science (DBCLS).
- Remove the liquid surrounding the fish to avoid lens effects.
- Measure the distance from the mouth to the point at which the tail begins.
Note: Juvenile zebrafish typically measure 12.5 mm. - Place the fish in the tank corresponding to its size.
- Fish that are too small can be harvested later.
- Fish that are too large can be used for later stages or breeding/stock maintenance.
- Fish that are too small can be harvested later.
- Check if the fish recovers fully from anesthesia.
After 3-5 min, the fish should start swimming normally again. Swimming may be sluggish at first, but the fish should recover fully within 5-10 min.
- Prepare the anesthetic solution: 200 ml of 1x MS222 solution, by diluting 10x MS222 solution (see Recipe 1) 1:9 in fish water in a fish tank (anesthesia tank).
- Zebrafish brain dissection
- Preparation of specimens and solutions
- Under the fume hood, prepare the fixing solution (4% PFA, 1x PBS, 0.1% Tween 20, see Recipe 2). Prepare 40 ml of fixing solution in a 50 ml conical tube for the prefixation of 5 fish heads. Keep the tubes at 4 °C.
- Kill the specimens in accordance with local regulations (e.g., European Union Directive 2010/63/EU, as developed in the recommendations of the Karlsruhe Institute of Technology for ‘The Humane Killing of Zebrafish’).
- Under the fume hood, prepare the fixing solution (4% PFA, 1x PBS, 0.1% Tween 20, see Recipe 2). Prepare 40 ml of fixing solution in a 50 ml conical tube for the prefixation of 5 fish heads. Keep the tubes at 4 °C.
- Prefixation of heads
- Transfer specimens to 1x PBS and cut the head behind the gills (keep the operculum intact), with a sharp blade to avoid any compression or tearing of the CNS.
- Place the head in a 50 ml conical tube containing fixing solution (transfer with tweezers).
- Incubate at 4 °C for 18 h.
- Transfer specimens to 1x PBS and cut the head behind the gills (keep the operculum intact), with a sharp blade to avoid any compression or tearing of the CNS.
- Dissection
- Under the fume hood, prepare fresh fixing solution. Prepare 40 ml of fixing solution in a 50 ml conical tube for 5 fish brains. Keep the tubes at 4 °C.
- Use tweezers to transfer a prefixed head to an SYLGARD-coated Petri dish (see Recipe 3) filled with PBST (see Recipe 4).
- Under a stereomicroscope, carefully extract the brain from head with fine tweezers. This step requires fine motor skills and it may take some training and practice to develop a satisfactory dissection technique. (see Supplemental Video)
- Remove the surrounding tissues from the brain with ultra-fine tweezers.
Notes:- Avoid touching the brain with the tweezers by using the connective tissues as handles.
- Avoid tearing the brain by applying only tangential forces while peeling away connective tissues.
- Avoid touching the brain with the tweezers by using the connective tissues as handles.
- Use an embryo spoon to transfer the dissected brain to the 50 ml conical tube containing fixing solution.
- Incubate at 4 °C for 5 h.
- Proceed to the next step of hydrogel incubation or keep the brain in storage solution.
- Under the fume hood, prepare fresh fixing solution. Prepare 40 ml of fixing solution in a 50 ml conical tube for 5 fish brains. Keep the tubes at 4 °C.
Note: For a detailed description of the dissection process we provide a dissection movie (see Supplemental Video). - Preparation of specimens and solutions
- Tissue clearing
- Incubation in hydrogel solution
- Under the fume hood, prepare the hydrogel solution on ice (see Recipe 5).
Note: The hydrogel solution must be prepared fresh, immediately before sample transfer into the solution. - Keep the samples on ice throughout the experiment.
- Transfer the samples into a 15 ml Falcon tube with a fine paintbrush or a cut transfer pipette.
Maximum: 8 brains/tube - Remove as much liquid as possible from the 15 ml tube.
- Add 14 ml of hydrogel solution to the 15 ml tube containing the samples.
Incubate at 4 °C for 48 h.
- Under the fume hood, prepare the hydrogel solution on ice (see Recipe 5).
- Polymerization
- Under the fume hood, set up the desiccator and set the hotplate to 37 °C.
- Label a 60 mm Petri dish with the name and number of the sample to be treated.
- Place the Petri dish on the 37 °C hotplate.
- Pour all 14 ml of hydrogel solution containing the brains into the Petri dish.
- Keep the Petri dish open and uncovered.
- Close the desiccator, ensuring a hermetic fit.
- Remove air with the vacuum pump until the pressure gauge reads -50 cm Hg.
- Carefully refill with pure nitrogen until the pressure gauge reads 0.
- Repeat the last two steps 6 times.
- Allow the samples to polymerize for 2.5 h.
- Under the fume hood, set up the desiccator and set the hotplate to 37 °C.
- Passive clearing in SDS solution
- Follow these steps after polymerization for brains:
- Place clean cassettes in a 60 mm Petri dish containing 8% SDS solution (see Recipe 6).
- Wearing multipolymer protective gloves, use curved tweezers to remove each brain individually from the polymerized hydrogel and place it in your hand.
- Using a fine, soft paintbrush and 8% SDS solution, gently clean the brain, removing all of the surrounding hydrogel.
- Place the brains in the cassettes.
- Close the cassettes and place them in a 300 ml beaker containing 8% SDS solution.
- Place clean cassettes in a 60 mm Petri dish containing 8% SDS solution (see Recipe 6).
- Cover the beaker with Parafilm and protect it from light for 2-5 h at room temperature, to dialyze out the remaining PFA, initiator and monomer.
- Place the cassettes in autoclaved rotisserie hybridization oven glass tubes containing 8% SDS solution.
- Incubate the hybridization tubes at 37 °C, with rotation, in the hybridization oven.
- Check the transparency of the samples every day (Figure 2).
Note: The standard time for clearing is 8 days for juvenile brains. - Once the sample has been successfully cleared (Figure 2), wash it three times in PBST. Replace the washing solution once daily for two more days.
- If the samples are not destined for use immediately after the third day of washing, store them in storage solution (see Recipe 8) at 4 °C.
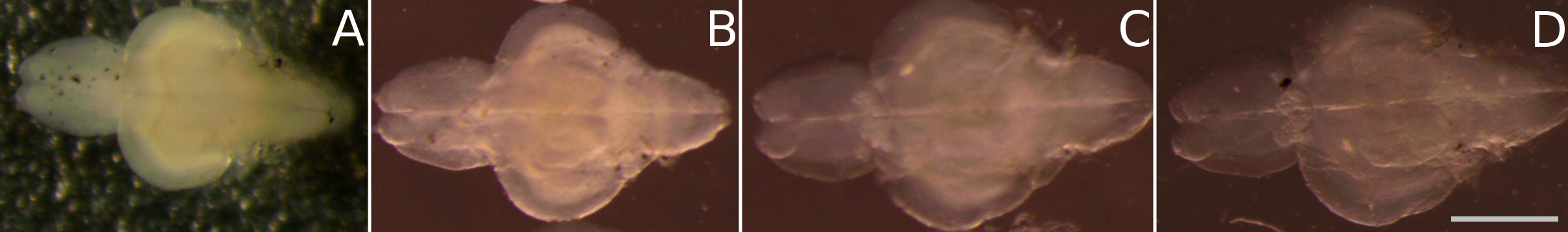
Figure 2. Time-series of the tissue clearing process: passive clearing of juvenile zebrafish brains (as described in steps C3c-C3e) is achieved within 8 days. This figure illustrates the clearing progress of juvenile fish brains in 8% SDS over time, showing an increase in transparency from: A. day 0, B. day 2, C. day 6, D. day 8. Scale bar = 1 mm.
- Follow these steps after polymerization for brains:
- Incubation in hydrogel solution
- Depigmentation
- Pour 10 ml of pre-incubation solution (SSC 0.5x-Tween 20, see Recipe 7) in a 60 mm Petri dish and transfer the samples to the Petri dish with a paintbrush or embryo spoon.
Note: Make sure you limit the dilution of the pre-incubation solution with PBST. - Incubate at room temperature for 1 h.
- Under the fume hood, prepare the depigmentation solution (see Recipe 7) immediately before use.
- Again under the fume hood, remove the samples with some liquid (to ensure that they don’t dry out) and place them in the lid of the Petri dish. Remove all of the pre-incubation solution from the dish and replace it with depigmentation solution. Return the samples to the dish with a minimum of pre-incubation solution.
Note: Keep the Petri dish uncovered and under light to accelerate the depigmentation process. - Check the progress of the depigmentation every 10 min. Cover the Petri dish when you check the depigmentation progress under a stereomicroscope.
Note: The standard depigmentation time is 45 min for juvenile brains. - Once the depigmentation is complete, rinse twice sequentially in PBST under a fume hood.
Note: Leave the rinsed samples on the bench overnight or for a minimum of 4 h before proceeding with the next step.
- Pour 10 ml of pre-incubation solution (SSC 0.5x-Tween 20, see Recipe 7) in a 60 mm Petri dish and transfer the samples to the Petri dish with a paintbrush or embryo spoon.
- Post-fixation
- Place the samples in a 2 ml Eppendorf tube. Remove all the liquid and add 1 ml of fixing solution.
- Incubate overnight at 4 °C.
Notes:- If the samples are to be used immediately after this step, rinse them four times in PBST for 3 h.
- If the samples are not going to be used immediately, remove the fixing solution and replace it with 1 ml of storage solution. Store the samples at 4 °C until use.
- If the samples are to be used immediately after this step, rinse them four times in PBST for 3 h.
- Place the samples in a 2 ml Eppendorf tube. Remove all the liquid and add 1 ml of fixing solution.
- Whole-mount labeling
- Transfer the samples to 2 ml glass tubes with a cut transfer pipette. Maximum: 5 brains/tube.
- To prevent dilution of the next solution, remove as much liquid as possible from the tube.
- Add 1 ml of blocking solution (see Recipe 9) to each tube.
- Incubate overnight at 4 °C.
- Remove the blocking solution and add 600 µl of staining solution (see Recipe 10) per tube.
- Add 1.2 µl of anti-GFP antibody per tube, to obtain a 1/500 dilution.
Note: Other primary antibodies have been successfully tested. But the dilution must be optimized, primary antibody concentration generally ranges from 1 µg/ml to 10 µg/ml. - Incubate at room temperature for 7 days, with gentle agitation, on a 3D rocker.
- Rinse 3 times, for 20 min each, in PBST.
- Remove all the liquid from the tube and add 600 µl of staining solution per tube.
- Add 1.5 µl of Alexa Fluor 488-conjugated secondary antibody per tube, to obtain a 1/400 dilution.
- Incubate at room temperature for 5 days, with gentle shaking, on a 3D rocker.
- Rinse for 2 days in several changes of PBST.
- Optional: DiI staining is used as a fiducial marker. Incubate the samples for 2 days in 600 µl of staining solution per tube at 1 µM dilution.
Note: Prepare 1 mM Dil stock solution by dissolving DiI powder in DMSO. - Rinse the DiI-stained sample in PBST for a minimum of 3 h.
- Optional: DiI staining is used as a fiducial marker. Incubate the samples for 2 days in 600 µl of staining solution per tube at 1 µM dilution.
- Transfer the samples to 2 ml glass tubes with a cut transfer pipette. Maximum: 5 brains/tube.
- Mounting and imaging
- Preparation for imaging
- Incubate the samples in 50% fbHRI/50% diluent for at least 6 h (see Recipe 11 and Figure 3B)
- Incubate in fbHRI for at least 12 h (Figure 3C).
- Incubate the samples in 50% fbHRI/50% diluent for at least 6 h (see Recipe 11 and Figure 3B)
- For imaging, mount samples in a droplet of 1% (wt/vol) low-melting point agarose in a 60 mm Petri dish, and cover them generously (to a depth of about 10 mm) with fbHRI.
- To avoid evaporation during prolonged imaging, cover with a layer of mineral oil.
- Whole-mount brain fluorescence can be imaged with a Leica TCS SP8 laser scanning confocal microscope equipped with a Leica HC FLUOTAR L 25x/1.00 IMM motCorr objective. The fluorescence signal is detected by exciting the fluorophores with lasers at wavelength of 488 and 552 nm, at a laser power of 1-3%, with capture by two internal photomultipliers (PMT). Tile-scan imaging followed by mosaic stitching is required to capture images of the whole brain.
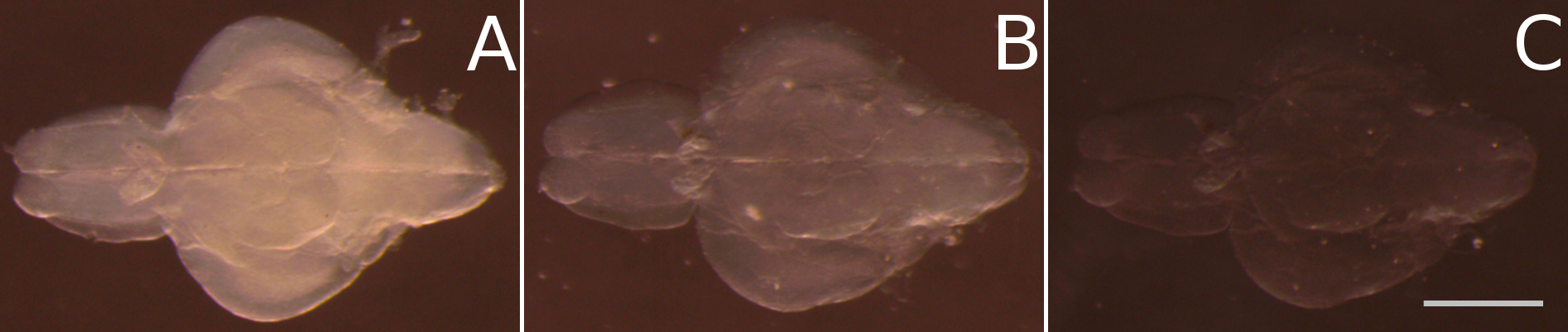
Figure 3. Final clearing of juvenile zebrafish brains after passive clearing by homogenizing the refractive index in fructose based high refractive index medium (fbHRI). A. After replacing the SDS solution of the passive clearing process with PBS the tissue turns opaque again. B. After incubation in 50% fbHRI in PBS the opacity is significantly reduced. C. After incubation in 100% fbHRI, the tissue clearing process is finished. Scale bar = 1 mm.
- Preparation for imaging
Data analysis
The analysis of imaging data depends strongly on the scientific questions addressed. Many methods are therefore available. The tissue-clearing techniques described here were used by Dambroise et al. (2017) to visualize maximum intensity projections of parts of complete specimens in Fiji. Manual and machine-assisted segmentations of expression patterns, and visualization of the resulting areas can be achieved in ITK-snap or Amira (FEI). Typical large specimens treated with the tissue clarification technique described here can be analyzed as shown in Video 1, a translation movie, in which each slice of the original stack is converted into a movie frame. Standard mp4-compression reduced the file size by several orders of magnitude, facilitating access.
Recipes
- 10x MS222 solution

Notes:- Filter solution (0.22 µm-pore filter).
- Store at room temperature.
- Filter solution (0.22 µm-pore filter).
- Fixing solution (4% PFA, 1x PBS, 0.1% Tween 20)

Notes:- Prepare under a fume hood.
- Use fresh.
- Store at 4 °C.
- Prepare under a fume hood.
- Sylgard-coated Petri dish
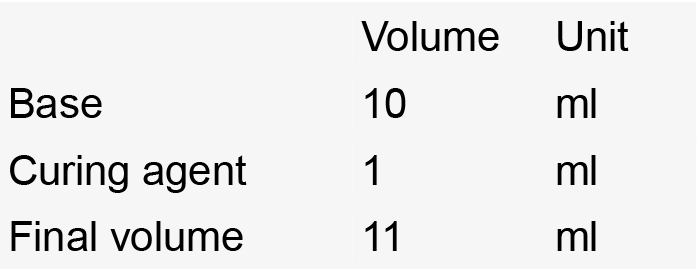
- Stir until homogeneous
- Degas
- Pour about 3 mm into a 60 mm-diameter Petri dish
- It will take up to 24 h to cure completely
- Stir until homogeneous
- PBST (1x PBS, 0.01% Tween 20)

Notes:- Filter solution (0.22 µm-pore filter).
- Store at room temperature.
- Filter solution (0.22 µm-pore filter).
- Hydrogel solution
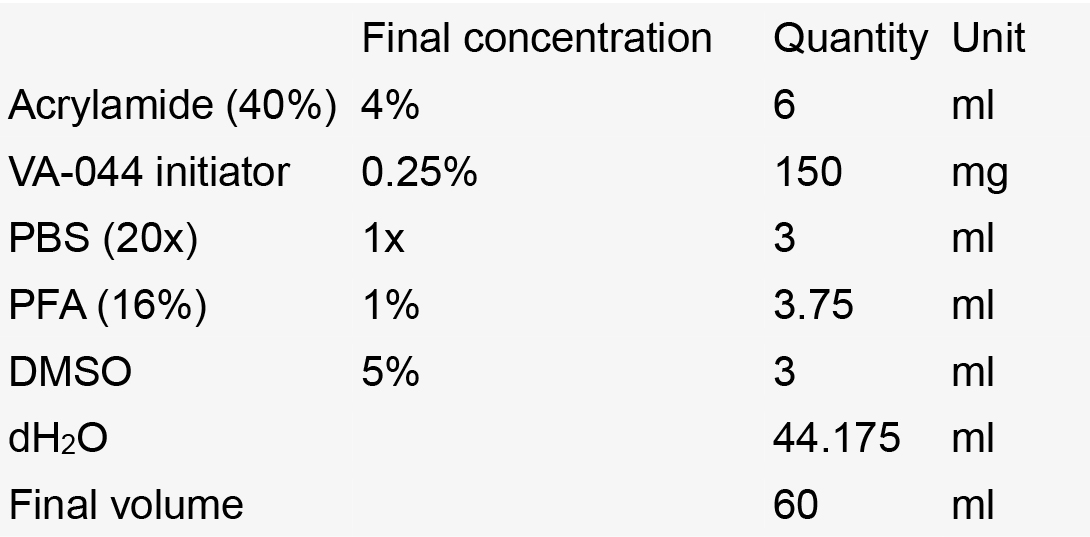
Notes:- Always make fresh.
- Prepare under the fume hood.
- Store at 4 °C.
- Always make fresh.
- SDS solution (8% SDS, 0.2 M boric acid, pH 8.5)

Notes:- Filter solution (0.22 µm-pore filter).
- Store at room temperature.
- Filter solution (0.22 µm-pore filter).
- Depigmentation
- Pre-incubation solution (SSC 0.5x-Tween 20)

- Depigmentation solution

Notes:- Always make fresh.
- Prepare under the fume hood.
- Always make fresh.
- Pre-incubation solution (SSC 0.5x-Tween 20)
- Storage solution
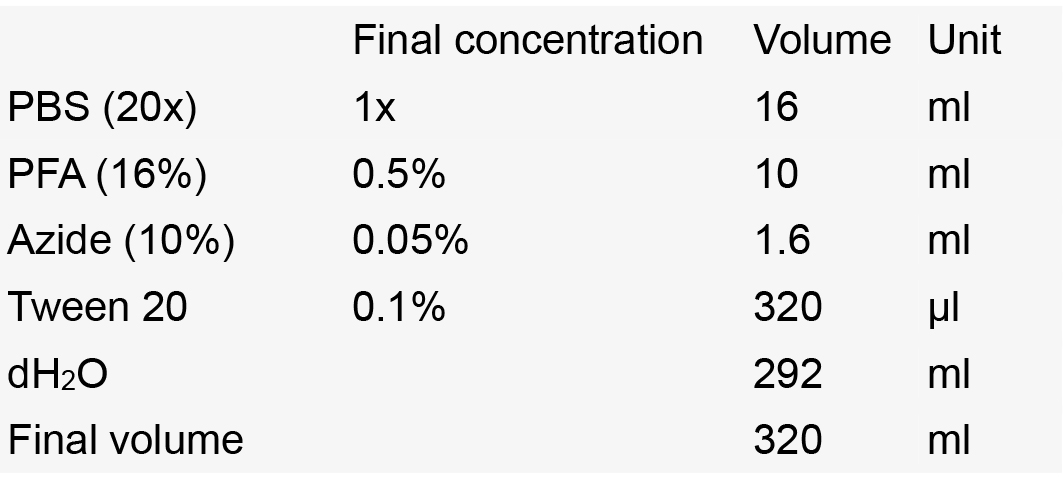
Notes:- Store at -20 °C.
- Keep at 4 °C after thawing.
- Store at -20 °C.
- Blocking solution
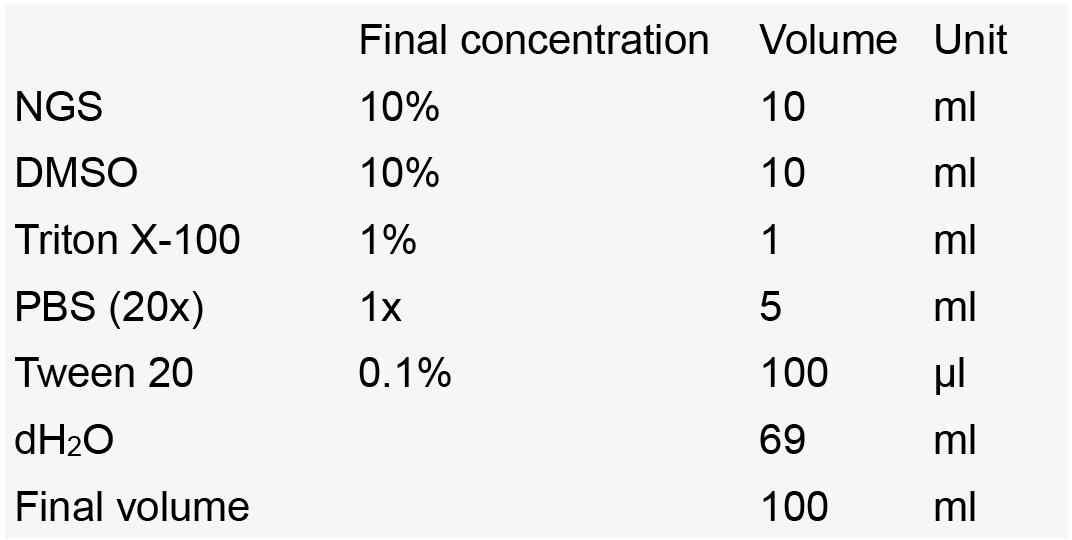
Notes:- Make aliquots of 10 ml in 15 ml Falcon tubes.
- Store at -20 °C.
- Keep at 4 °C after thawing.
- Make aliquots of 10 ml in 15 ml Falcon tubes.
- Staining solution
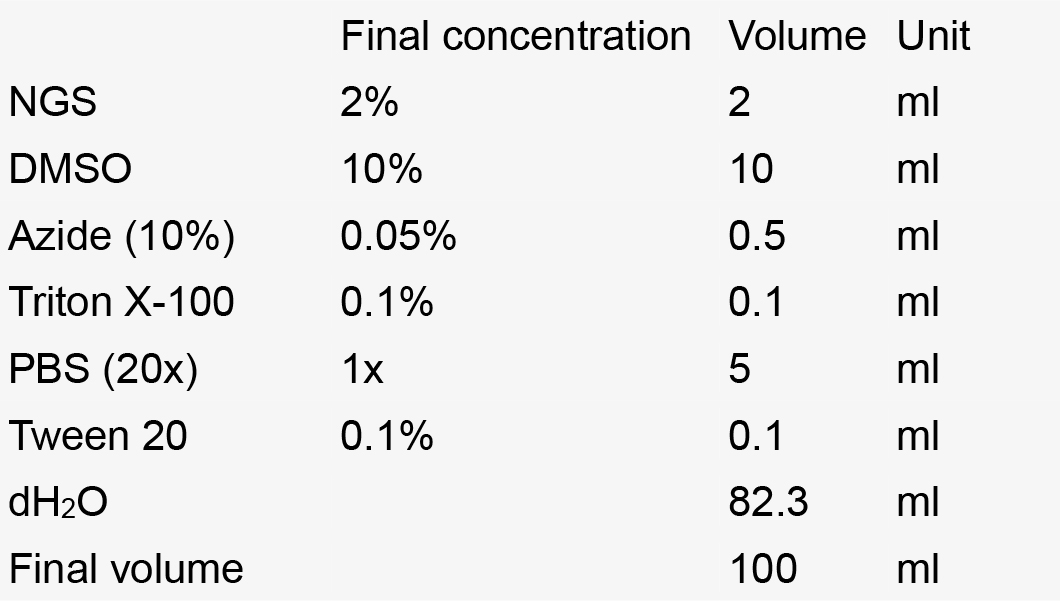
Notes:- Make aliquots of 10 ml in 15 ml Falcon tubes.
- Store at -20 °C.
- Keep at 4 °C after thawing.
- Make aliquots of 10 ml in 15 ml Falcon tubes.
- Fructose-based high-refractive index solution (fbHRI)
The fbHRI solution is generated by mixing three solutions prepared separately in advance:- Aqueous diluent with a refractive index (RI) of ~1.334

Notes:- No need to adjust RI.
- Store at 4 °C.
- No need to adjust RI.
- ~90% fructose solution with an RI of 1.457 (F1457)

- In a beaker, dissolve 800 g of fructose in ~ 300 ml diluent, mix with a spoon
- Cover the fructose solution to prevent evaporation and stir with a strong magnetic stirrer overnight to obtain a homogenous solution. Make up the volume to 800 ml with diluent to obtain a 100% (w/v) fructose solution. Set aside 50 ml of 100% (w/v) fructose for final RI adjustment
- Measure RI and adjust the fructose solution to an RI of 1.457 with diluent
- In a beaker, dissolve 800 g of fructose in ~ 300 ml diluent, mix with a spoon
- ~85% DMSO solution with an RI of 1.457 (DMSO1457)

Measure RI and adjust the DMSO solution to an RI of 1.457 with diluent - fbHRI solution
- Mix F1457 and DMSO1457 in the ratio of 80/20
- Measure refractive index with a refractometer
- Adjust RI:
Add 100% fructose to raise RI, add diluent to lower RI - Aliquot into 50 ml Falcon tubes and store at 4 °C
- Control and adjust refractive index to 1.457 with a refractometer before use
- Mix F1457 and DMSO1457 in the ratio of 80/20
- Aqueous diluent with a refractive index (RI) of ~1.334
Acknowledgments
We would like to thank Dr. Spencer Brown for suggesting the use of DiIC18 as a fiducial marker. This work benefited from the facilities and expertise of TEFOR-Investissement d’avenir, and support from the Association Nationale de la Recherche et de la Technologie (ANR-II-INBS-0014). The authors declare no competing financial interests.
References
- Ampatzis, K., Makantasi, P. and Dermon, C. R. (2012). Cell proliferation pattern in adult zebrafish forebrain is sexually dimorphic. Neuroscience 15: 367-381.
- Bailey, J. M., Oliveri, A. N. and Levin, E. D. (2015). Pharmacological analyses of learning and memory in zebrafish (Danio rerio). Pharmacol Biochem Behav 139 Pt B: 103-111.
- Chung, K., Wallace, J., Kim, S. Y., Kalyanasundaram, S., Andalman, A. S., Davidson, T. J., Mirzabekov, J. J., Zalocusky, K. A., Mattis, J., Denisin, A. K., Pak, S., Bernstein, H., Ramakrishnan, C., Grosenick, L., Gradinaru, V. and Deisseroth, K. (2013). Structural and molecular interrogation of intact biological systems. Nature 497(7449): 332-337.
- Dambroise, E., Simion, M., Bourquard, T., Bouffard, S., Rizzi, B., Jaszczyszyn, Y., Bourge, M., Affaticati, P., Heuze, A., Jouralet, J., Edouard, J., Brown, S., Thermes, C., Poupon, A., Reiter, E., Sohm, F., Bourrat, F. and Joly, J. S. (2017). Postembryonic fish brain proliferation zones exhibit neuroepithelial-type gene expression profile. Stem Cells 35(6): 1505-1518.
- Filippi, A., Mahler, J., Schweitzer, J., Driever, W. J. (2010). Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. J Comp Neurol 518(4): 423-38.
- Fini, J. B., Mughal, B. B., Le Mevel, S., Leemans, M., Lettmann, M., Spirhanzlova, P., Affaticati, P., Jenett, A. and Demeneix, B. A. (2017). Human amniotic fluid contaminants alter thyroid hormone signalling and early brain development in Xenopus embryos. Sci Rep 7: 43786.
- Frétaud, M., Riviere, L., Job, E., Gay, S., Lareyre, J. J., Joly, J. S., Affaticati, P. and Thermes, V. (2017). High-resolution 3D imaging of whole organ after clearing: taking a new look at the zebrafish testis. Sci Rep 7: 43012.
- Ke, M. T., Fujimoto, S. and Imai, T. (2013). SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16(8): 1154-1161.
- Mayrhofer, M., Gourain, V., Reischl, M., Affaticati, P., Jenett, A., Joly, J. S., Benelli, M., Demichelis, F., Poliani, P. L., Sieger, D. and Mione, M. (2017). A novel brain tumour model in zebrafish reveals the role of YAP activation in MAPK- and PI3K-induced malignant growth. Dis Model Mech 10(1): 15-28.
- Lawrence, C., Best, J., Cockington, J., Henry, E. C., Hurley, S., James, A., Lapointe, C., Maloney, K. and Sanders, E. (2016). The complete and updated “Rotifer Polyculture Method” for rearing first feeding zebrafish. J Vis Exp (107): e53629.
- Lawrence, C. and Mason, T. (2012). Zebrafish housing systems: a review of basic operating principles and considerations for design and functionality. ILAR J 53(2): 179-191.
- Oksanen, K. E., Halfpenny, N. J., Sherwood, E., Harjula, S. K., Hammaren, M. M., Ahava, M. J., Pajula, E. T., Lahtinen, M. J., Parikka, M. and Ramet, M. (2013). An adult zebrafish model for preclinical tuberculosis vaccine development. Vaccine 31(45): 5202-5209.
- Than-Trong, E. and Bally-Cuif, L. (2015). Radial glia and neural progenitors in the adult zebrafish central nervous system. Glia 63(8): 1406-1428.
- Tomer, R., Ye, L., Hsueh, B. and Deisseroth, K. (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc 9(7): 1682-1697.
- Treweek, J. B., Chan, K. Y., Flytzanis, N. C., Yang, B., Deverman, B. E., Greenbaum, A., Lignell, A., Xiao, C., Cai, L., Ladinsky, M. S., Bjorkman, P. J., Fowlkes, C. C. and Gradinaru, V. (2015). Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc 10(11): 1860-1896.
- Xavier, A. L., Fontaine, R., Bloch, S., Affaticati, P., Jenett, A., Demarque, M., Vernier, P. and Yamamoto, K. (2017). Comparative analysis of monoaminergic cerebrospinal fluid-contacting cells in Osteichthyes (bony vertebrates). J Comp Neurol 525(9): 2265-2283.
- Yang, B., Treweek, J. B., Kulkarni, R. P., Deverman, B. E., Chen, C. K., Lubeck, E., Shah, S., Cai, L. and Gradinaru, V. (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158(4): 945-958.
- Parichy DM, Elizondo MR, Mills MG, Gordon TN and Engeszer RE. (2009). Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn 238(12): 2975-3015.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Affaticati, P., Simion, M., De Job, E., Rivière, L., Hermel, J., Machado, E., Joly, J. and Jenett, A. (2017). zPACT: Tissue Clearing and Immunohistochemistry on Juvenile Zebrafish Brain. Bio-protocol 7(23): e2636. DOI: 10.21769/BioProtoc.2636.
Category
Neuroscience > Neuroanatomy and circuitry > Animal model
Cell Biology > Tissue analysis > Tissue staining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












