- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation, BODIPY Labeling and Uptake of Exosomes in Hepatic Stellate Cells
Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2633 Views: 12239
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Assays for Measuring Endothelial Permeability by Transwells and Electrical Impedance Systems
Hong-Ru Chen and Trai-Ming Yeh
May 5, 2017 28044 Views

TZA, a Sensitive Reporter Cell-based Assay to Accurately and Rapidly Quantify Inducible, Replication-competent Latent HIV-1 from Resting CD4+ T Cells
Anwesha Sanyal [...] Phalguni Gupta
May 20, 2019 7794 Views

Generation and Implementation of Reporter BHK-21 Cells for Live Imaging of Flavivirus Infection
Jorge L. Arias-Arias and Rodrigo Mora-Rodríguez
Mar 5, 2021 4367 Views
Abstract
Exosomes have emerged as an important mediator of intercellular communication. They are present in extracellular milieu and therefore, easily accessible by neighboring or distant cells. They carry mRNA, microRNAs and proteins within their vesicles and once internalized by recipient cells; they can modulate multiple signaling pathways with pleiotropic effects from inducing antiviral state to disease progression. We have previously shown that hepatitis C virus (HCV) infected hepatocytes or hepatoma cells harboring genome-length replicon secrete exosomes in culture supernatants. These exosomes are taken up by hepatic stellate cells (HSC) and activate them to induce fibrosis during HCV infection. Here, we describe detailed protocols for exosomes isolation and uptake of BODIPY labeled exosomes by hepatic stellate cells.
Keywords: ExosomesBackground
Exosomes are small membrane-bound extracellular vesicles of 40-150 nm in diameter. They have been identified in all body fluids, including urine, amniotic fluid, serum, saliva, breast milk, cerebrospinal fluid, nasal secretions and in the supernatant of tissue cultured cell lines. Exosomes mediate cell to cell communication through transfer of microRNAs (miRs), mRNAs, and proteins, etc. which can be taken up by neighboring or distant cells and subsequently promote signaling in recipient cells (Schorey et al., 2015). Exosomes are enriched in proteins, including members of the tetraspanin family (CD9, CD81, CD63), heat shock proteins (Hsp60, Hsp70, Hsp90) and proteins of the multivesicular bodies (annexins, RabGTPases and endosomal sorting complexes required for transport [ESCRT] proteins). Recent studies suggest that exosomes can influence immune response (Li et al., 2013; Sun et al., 2016), facilitate productive infection in naïve hepatoma cells (Ramakrishnaiah et al., 2013; Shrivastava et al., 2015), disease progression (Devhare et al., 2017) as well as drug resistance to anti-cancer therapy (Qu et al., 2016). Most commonly used techniques for isolation of exosomes involves ultracentrifugation (UC) and ExoQuick (EQ) precipitation. In hepatitis C virus (HCV) infection, crosstalk between liver resident cells including hepatocytes, macrophages, endothelial cells, lymphocytes and stellate cells play an important role in disease progression. We have shown earlier that HCV infected hepatocytes or hepatoma cells harboring genome-length replicon secrete exosomes in the cell culture supernatants (Shrivastava et al., 2015). These exosomes carried miR-19a that induces fibrogenesis in hepatic stellate cells that might lead to liver fibrosis during HCV infection (Devhare et al., 2017). Here, we provide a protocol for labeling of exosomes with green fluorescent lipophilic dye, BODIPY that enables real-time monitoring and localization of exosomes within the recipient hepatic stellate cells.
Materials and Reagents
- Serological pipettes (5 ml, 10 ml, 25 ml) (Corning)
- 50 ml polypropylene centrifuge tubes (Corning, catalog number: 430829 )
- T-25 cm2 cell culture flask (Corning, catalog number: 430639 )
- 100 mm tissue culture treated culture dish (Corning, catalog number: 430167 )
- Ultra-clear centrifuge tubes, 13.2 ml, 14 x 89 mm for SW41Ti rotor (Beckman Coulter, catalog number: 344059 )
- 15 ml polypropylene centrifuge tubes (Corning, catalog number: 430791 )
- 4-well chambered slides (Thermo Fisher Scientific, Nunc, catalog number: 177437 )
- Aluminium foil (Fisher Scientific, FisherbrandTM, catalog number: 01-213-102 )
- 0.2 μm size filter (Acrodisc Syringe Filters) (Pall, catalog number: 4612 )
- Huh7.5 cell line (Human hepatocellular carcinoma cell line)
- Rep2a-Rluc cell line: Huh7.5 cells harboring genome-length replicon of hepatitis C virus
- LX2 cells (immortalized human hepatic stellate cells)
- DMEM medium (Sigma-Aldrich, catalog number: D5796 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 16000044 )
- Penicillin/streptomycin (Sigma-Aldrich, catalog number: P0781 )
- Phosphate buffered saline (DPBS) without CaCl2 and MgCl2 (Sigma-Aldrich, catalog number: D8537 )
- 0.25% trypsin-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25200056 )
- ExoQuick-TC (System Biosciences, catalog number: EXOTC10A-1 )
- Poly-L-lysine (Sigma-Aldrich, catalog number: P4707 )
- 4,6-Diamidino-2-phenylindole dihydrochloride(DAPI) (Sigma-Aldrich, catalog number: D9542 )
- Alexa Fluor-647 conjugated wheat germ agglutinin (WGA) (Thermo Fisher Scientific, InvitrogenTM, catalog number: W32466 )
- 4,4-Difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D3922 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650 )
- Formaldehyde solution (37 wt. % in H2O) (Sigma-Aldrich, catalog number: 252549 )
- TritonX-100 (Sigma-Aldrich, catalog number: T8787 )
- Hanks’ balanced salt solution (HBSS) (Sigma-Aldrich, catalog number: H6648 )
- BODIPY stock solution (see Recipes)
- Exosome depleted serum (see Recipes)
- Fixation solution (see Recipes)
- Permeabilization buffer (see Recipes)
- WGA conjugate stock solution (see Recipes)
Equipment
- Pipette-aid
- Water bath
- Centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: Sorvall ST 8R )
- 37 °C with 5% CO2 cell culture incubator
- Inverted microscope (Nikon Instruments, model: Eclipse TS100 , Objective: 10x)
- SW41Ti rotor (Beckman Coulter, model: SW41Ti )
- Zetasizer Nano (Malvern Instruments, model: NanoSight LM10 )
- Olympus FV1000 confocal microscope (Olympus, model: FV1000 , Objective:60x)
- Biosafety cabinet
- Ultracentrifuge (Beckman Coulter, model: L8-80M )
Procedure
- Revival of Huh7.5 or Rep2a-Rluc (hepatocytes) cell lines
- Rapidly thaw frozen Huh7.5 or Rep2a-Rluc cells (~1 ml) in a 37 °C water bath.
- Transfer entire content of Huh7.5 or Rep2a-Rluc cells into a sterile 50 ml conical tube and slowly add 10 ml of pre-warmed DMEM media containing 10% FBS and 100 U/ml of penicillin and streptomycin (complete media).
- Mix well, and centrifuge at 300 x g for 10 min at RT.
- Discard the supernatant and resuspend cell pellet in 5 ml of complete media and transfer to a T-25 cm2 culture flask.
- Incubate cells at 37 °C in a 5% CO2 incubator.
- Observe culture flasks routinely under inverted microscope using 10x objective and split cells one day before the experiment.
Note: Next day after revival, observe the culture flask for floating cells. If floating cells are seen, replace flask with fresh 5 ml of complete media. Hepatoma cells should be split in 1:3 to 1:5 ratio in every 3-5 days to maintain the cell growth.
- Rapidly thaw frozen Huh7.5 or Rep2a-Rluc cells (~1 ml) in a 37 °C water bath.
- BODIPY labeling of exosomes
- Seed 2 x 106 Huh7.5 cells of ~50% confluency in a 100 mm plate and incubate at 37 °C in 5% CO2 incubator overnight. Next day, infect cells with HCV at a multiplicity of infection (MOI) of 0.1. Allow virus to adsorb onto the cells for 8 h in a minimum volume (2 ml) of DMEM with 2% of FBS and antibiotics. Wash the virus-infected cells 3 times with PBS.
Note: Media with reduced serum concentration helps in virus infection. PBS should be kept at room temperature before use. - Alternatively, seed 3-4 x 106 Rep2a-Rluc cells of ~80% confluency in a 100 mm plate and incubate cells at 37 °C in 5% CO2 incubator overnight. Next day, wash the cells 3 times with PBS.
- Dilute 10 μl of 1 mM of BODIPY 493/503 solution (see Recipes) to 10 ml of culture media to make the final concentration of 1 μM, mix well and add onto the virus infected Huh7.5 or Rep2a-Rluc cells.
- Incubate cells for 1 h at 37 °C in 5% CO2 incubator.
- Wash the cells 3 times with PBS.
- Add 10 ml of fresh DMEM media with 2% of exosome depleted sera (see Recipes) onto the cells and incubate for either 3 days or 24 h for HCV infected Huh7.5 or Rep2a-Rluc cells respectively.
- After incubation, collect culture supernatant and proceed for exosomes isolation.
- Seed 2 x 106 Huh7.5 cells of ~50% confluency in a 100 mm plate and incubate at 37 °C in 5% CO2 incubator overnight. Next day, infect cells with HCV at a multiplicity of infection (MOI) of 0.1. Allow virus to adsorb onto the cells for 8 h in a minimum volume (2 ml) of DMEM with 2% of FBS and antibiotics. Wash the virus-infected cells 3 times with PBS.
- Exosome isolation by ultracentrifugation
- Collect 10 ml of culture supernatant from HCV infected Huh7.5 or Rep2a-Rluc cells and centrifuge at 300 x g at 4 °C for 5 min. Without disturbing the cell pellet, carefully transfer the supernatant to a new ultra-clear centrifuge tube.
- To get rid of possible cell debris, centrifuge the supernatant at 2,000 x g for 10 min at 4 °C, followed by 26,500 x g for 30 min at 4 °C using SW41 Ti rotor by ultracentrifugation. Transfer the supernatant to a new ultra-clear centrifuge tube.
- Then, centrifuge the supernatant using SW41 Ti rotor at 110,000 x g for 90 min at 4 °C by ultracentrifugation to isolate the exosomes. Discard the supernatant.
- Wash the exosome pellet 2 times with PBS. For this, resuspend pellet in 10 ml of PBS and centrifuge at 110,000 x g for 60 min at 4 °C. Discard the supernatant. Repeat this step again.
- Resuspend the final pellet referred to as exosomes in PBS to 1/20 of the original volume of culture supernatant for analysis.
- Check the size distribution of exosomes by dynamic light scattering (DLS) using a Zetasizer Nano (Malvern Instruments). Size distribution analysis of exosomes by DLS is shown in Figure 1. Exosome size is measured as diameter in nm (d. nm) and Polydispersity index (PDI) indicates size distribution of exosomes within the sample.
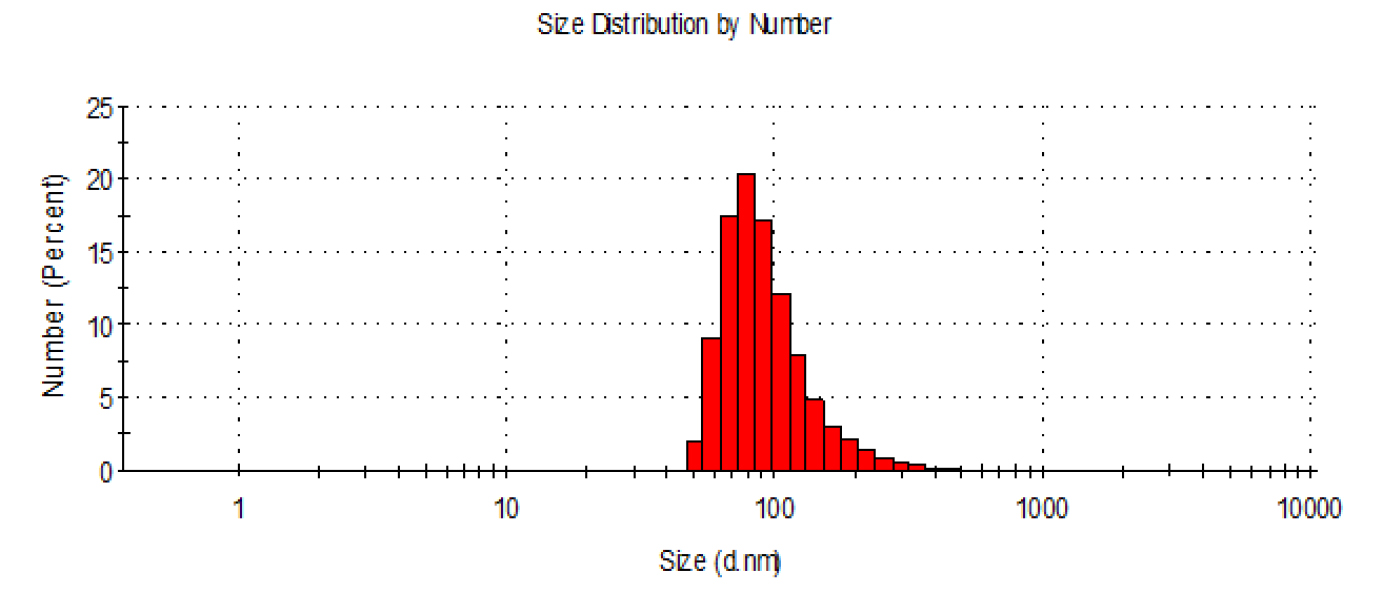 Figure 1. Size distribution analysis of purified exosomes by DLS (Nano-ZS90, Malvern). Mean value from the intensity distribution, Z average is 202 nm and polydispersity index, PDI is 0.34.
Figure 1. Size distribution analysis of purified exosomes by DLS (Nano-ZS90, Malvern). Mean value from the intensity distribution, Z average is 202 nm and polydispersity index, PDI is 0.34.
- Collect 10 ml of culture supernatant from HCV infected Huh7.5 or Rep2a-Rluc cells and centrifuge at 300 x g at 4 °C for 5 min. Without disturbing the cell pellet, carefully transfer the supernatant to a new ultra-clear centrifuge tube.
- Exosome isolation by ExoQuick method
- Collect 10 ml of culture supernatant from HCV infected Huh7.5 or Rep2a-Rluc cells.
- To get rid of possible cell debris, centrifuge at 3,000 x g at 4 °C for 15 min. Transfer the supernatant to a new 15 ml conical tube.
- Add ExoQuick at a ratio of 5:1 in the supernatant (5 ml of culture supernatant and 1 ml of ExoQuick), mix well and incubate overnight at 4 °C.
- Centrifuge the cell culture supernatant and ExoQuick mixture at 2,000 x g for 30 min at 4 °C.
- Remove the residual supernatant and resuspend the final pellet referred to as exosomes in PBS for analysis.
- Collect 10 ml of culture supernatant from HCV infected Huh7.5 or Rep2a-Rluc cells.
- Uptake of BODIPY labeled exosomes
- Grow immortalized human hepatic stellate cell lines, LX2 in DMEM media containing 10% FBS and 100 U/ml of antibiotics (complete media).
- Pre-coat the 4-well chamber slides with poly-L-lysine (0.01%) for 1 h at 37 °C.
- Split LX2 cells and resuspend cell pellet in DMEM media supplemented with 2% exosome depleted FBS and antibiotics.
- Seed 1 x 104 LX2 cells in each well of 4-well chamber slides.
- Incubate cells at 37 °C in 5% CO2 incubator overnight.
- Next day, expose LX2 cells with BODIPY labeled exosomes for 3 h (short) or 24 h (long).
Note: Exosomes should be diluted in serumfree media from 1:2 to 1:10 before adding onto LX2 cells. Do not add exosomes directly onto the cells. Since exosomes are resuspended in PBS, longer incubation of cells with exosomes will detach cells from chamber slides. To observe any changes at molecular level such as gene expression, exosomes should be incubated with recipient cells for a minimum of 24 h. - After incubation, process LX2 cells for confocal microscopy to observe exosome internalization.
- Grow immortalized human hepatic stellate cell lines, LX2 in DMEM media containing 10% FBS and 100 U/ml of antibiotics (complete media).
- Staining of cells for confocal microscopy
- Fix the cells with fixation solution (3.7% formaldehyde, see Recipes) for 20 min at RT.
- Wash the cells 3 times with PBS.
- Permeabilize cells with permeabilization buffer (0.2% TritonX-100, see Recipes) for 5 min at RT.
- Wash the cells 3 times with PBS.
- Stain the cell nuclei with DAPI (1 µg/ml) for 2 min at RT (blue color) and observed under a confocal microscope. A representative image is shown in Figure 2A.
- For cell boundary or plasma membrane staining, incubate live LX2 cells with 5 µg/ml of Alexa Fluor-647 conjugated WGA (see Recipes) for 10 min at 37 °C.
- Wash the cells 2 times with HBSS.
- Fix the cells and follow the procedure as mentioned above. A representative image is shown in Figure 2B.
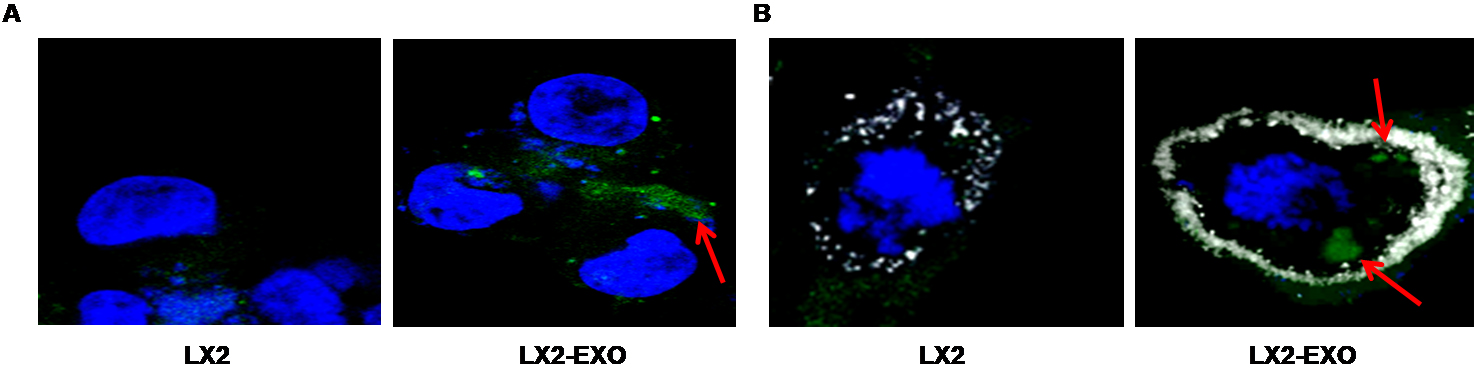
Figure 2. Internalization of BODIPY labeled exosomes by hepatic stellate cells (LX2). BODIPY labeled exosomes are isolated from the culture supernatants of HCV infected hepatocytes. LX2 cells are exposed with BODIPY labeled exosomes for 3 h. A. Immunofluorescence images showing uptake of BODIPY labeled exosomes (green color) and nuclei (blue color) by LX2 cells. B. Immunofluorescence images showing uptake of BODIPY labeled exosomes (green color), WGA (white color) to mark cell boundary and nuclei (blue color) by LX2 cells. The red arrow indicates BODIPY labeled exosomes inside the cell. The images are taken at 60x2 X magnification.
Notes:- It is very hard to observe evenness of exosome labeling by microscopy. They are too small to be individually resolved. Generally, under microscope, BODIPY labeled exosomes tend to clump together. Exosomes size vary from 50-150 nm in diameter, however, high protein content in exosomes increases the size up to 200 nm in diameter.
- Two commonly used method for exosome isolation worked on different principles. Ultracentrifugation involves differential centrifugation. Successive rounds of centrifugation are intended to pellet down sequentially apoptotic bodies, cell debris, shedding vesicles and then, the exosomes are isolated based on size and density. It is a timeconsuming process involves multiple steps and potentially lead to exosomal aggregation. Exoquick method involves polymer based precipitation technique. There is a chance of contamination of exosomes with microvesicles and apoptotic bodies, however, exosomes can be isolated from very small amount of sample by this method.
- It is very hard to observe evenness of exosome labeling by microscopy. They are too small to be individually resolved. Generally, under microscope, BODIPY labeled exosomes tend to clump together. Exosomes size vary from 50-150 nm in diameter, however, high protein content in exosomes increases the size up to 200 nm in diameter.
- Fix the cells with fixation solution (3.7% formaldehyde, see Recipes) for 20 min at RT.
Data analysis
Each experiment has been repeated at least three times to verify reproducibility.
Recipes
- BODIPY stock solution
- Prepare 5 mM (1.25 mg/ml) stock solution in DMSO
- Store the stock solution at -20 °C wrapped in aluminum foil
- Protect it from light and make smaller aliquots to avoid repeated freeze-thaw
- Prepare 1 mM working solution in DMSO at the time of experiment
- Prepare 5 mM (1.25 mg/ml) stock solution in DMSO
- Exosome depleted serum
Ultracentrifuge FBS at 110,000 x g at 4 °C for 16 h using SW41 Ti rotor (Beckman Coulter), collect the supernatant and then pass through a 0.22 µm filter
Store the filtered exosome depleted serum at 4 °C for further use - Fixation solution
Prepare 3.7% formaldehyde by diluting 37% formaldehyde solution in PBS - Permeabilization buffer
Add 2 µl of Triton X-100 in 1 ml of PBS
Always prepare fresh buffer at the time of experiment and vortex it properly - WGA conjugate stock solution
- Prepare 1 mg/ml stock solution by dissolving 5 mg of lyophilized powder in 5 ml of HBSS
- Store the stock solution at -20 °C wrapped in aluminum foil
- Protect it from light and make smaller aliquots to avoid repeated freeze-thaw
- Prepare 1 mg/ml stock solution by dissolving 5 mg of lyophilized powder in 5 ml of HBSS
Acknowledgments
This protocol is adapted from previously published paper (Devhare et al., 2017). Huh7.5, Rep2a-Rluc and LX2 cell lines are kindly provided by Charles M Rice, Rockefeller University, New York, USA, Hengli Tang, Florida State University, Florida, USA and Scott Friedman, Mount Sinai School of Medicine, NY, USA respectively. The author declares that there is no conflict of interest.
References
- Devhare, P. B., Sasaki, R., Shrivastava, S., Di Bisceglie, A. M., Ray, R. and Ray, R. B. (2017). Exosome-mediated intercellular communication between hepatitis C virus-infected hepatocytes and hepatic stellate cells. J Virol 91(6): e02225.
- Li, J., Liu, K., Liu, Y., Xu, Y., Zhang, F., Yang, H., Liu, J., Pan, T., Chen, J., Wu, M., Zhou, X. and Yuan, Z. (2013). Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat Immunol 14(8): 793-803.
- Qu, Z., Wu, J., Wu, J., Luo, D., Jiang, C. and Ding, Y. (2016). Exosomes derived from HCC cells induce sorafenib resistance in hepatocellular carcinoma both in vivo and in vitro. J Exp Clin Cancer Res 35(1): 159.
- Ramakrishnaiah, V., Thumann, C., Fofana, I., Habersetzer, F., Pan, Q., de Ruiter, P. E., Willemsen, R., Demmers, J. A., Stalin Raj, V., Jenster, G., Kwekkeboom, J., Tilanus, H. W., Haagmans, B. L., Baumert, T. F. and van der Laan, L. J. (2013). Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110(32): 13109-13113.
- Schorey, J. S., Cheng, Y., Singh, P. P. and Smith, V. L. (2015). Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep 16(1): 24-43.
- Shrivastava, S., Devhare, P., Sujijantarat, N., Steele, R., Kwon, Y. C., Ray, R. and Ray, R. B. (2015). Knockdown of autophagy inhibits infectious hepatitis C virus release by the exosomal pathway. J Virol 90(3): 1387-1396.
- Sun, L., Wang, X., Zhou, Y., Zhou, R. H., Ho, W. Z. and Li, J. L. (2016). Exosomes contribute to the transmission of anti-HIV activity from TLR3-activated brain microvascular endothelial cells to macrophages. Antiviral Res 134: 167-171.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shrivastava, S. (2017). Isolation, BODIPY Labeling and Uptake of Exosomes in Hepatic Stellate Cells. Bio-protocol 7(23): e2633. DOI: 10.21769/BioProtoc.2633.
Category
Microbiology > Microbial biochemistry > Lipid
Microbiology > Microbial cell biology > Cell-based analysis > Reporter assay
Biochemistry > Lipid > Extracellular lipids
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










