- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Mn Concentrations in Synechocystis sp. PCC6803 Using ICP-MS
(*contributed equally to this work) Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2623 Views: 8588
Reviewed by: Maria SinetovaElizabeth LibbyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1766 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1810 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1781 Views
Abstract
Manganese (Mn) is an essential micronutrient for all photoautotrophic organisms. Two distinct pools of Mn have been identified in the cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis), with 80% of the Mn residing in the periplasm and 20% in cytoplasm and thylakoid lumen (Keren et al., 2002). In this protocol, we describe a method to quantify the periplasmic and intracellular pools of Mn in Synechocystis accurately, using inductively coupled plasma mass spectrometry (ICP-MS).
Keywords: CyanobacteriaBackground
Mn plays a vital role in the active sites of several enzymes such as the oxygen-evolving complex in photosystem II. In contrast to its role as an important micronutrient, Mn can be toxic when present in excess. It is therefore of crucial importance for cyanobacteria to maintain the intracellular levels of Mn and in particular to avoid free Mn in the cytoplasm. The cyanobacterium Synechocystis addresses this challenge by storing about 80% of the Mn in the periplasm. Only 20% of the cellular content can be detected in the cytoplasm and thylakoid system (Keren et al., 2002), with most of the Mn being incorporated into proteins, leaving virtually no free Mn in the cytoplasm. We recently identified the manganese transport protein Mnx, which resides in the thylakoid membrane and facilitates export of Mn from the cytoplasm into the thylakoid lumen in Synechocystis. According to our study, Mnx is a major player in maintaining the cellular Mn homeostasis (Brandenburg et al., 2017). To analyze the biological significance of Mnx, we developed a protocol to measure the periplasmic and intracellular Mn pools separately. We chose ICP-MS for quantification, since it is a sensitive and reliable method to detect metals in biological samples. Detection limits can be in the range of [ng L-1] and below. Prior to the analysis, all complex molecules of the sample are broken down to atomic compounds by digestion with nitric acid. Subsequently, the sample is ionized by an inductively coupled plasma and analyzed by mass spectrometry. In this protocol, we describe the detailed workflow for subcellular Mn quantification, from sampling to the calculation of Mn concentrations.
Materials and Reagents
- 15 ml tubes (SARSTEDT, catalog number: 62.554.002 )
- Syringe filter filtropur S, 0.2 µm (SARSTEDT, catalog number: 83.1826.001 )
- 10 ml Luer-Lok syringe without needle (BD, catalog number: 300912 )
- Milli-Q grade (or similar) water (Resistivity at 25 °C 18 MΩ cm)
- Distilled AR grade nitric acid (Avantor Performance Materials, MACRON, catalog number: 1409-46 )
- ICP-MS multi element standard solution VI (Merck, catalog number: 1105800100 )
- Na2-Ethylenediaminetetraacetic acid (EDTA) (Carl Roth, catalog number: 8043.2 )
- 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Carl Roth, catalog number: 6763.3 )
- EDTA wash buffer (see Recipes)
Equipment
- Cooling centrifuge for 15 ml tubes (Beckman Coulter, model: GS-6 )
- Teflon cups (Savillex, custom made)
- Analytical balance (KERN & SOHN, model: ALJ 220-4M )
- Vortex Mixer (Eppendorf, model: MixMate® )
- Clean lab Class 5000 (according to US FED STD 209E, which would be in between ISO6 and ISO 7 according to the newer ISO 14644-1 and ISO 14698 standards)
- Chemical hood (Wesemann)
- Hot plate (Ceramic Stirring Hot Plate) (IKA, catalog number: 35810.01 )
- ICP-MS 7500cx (Agilent Technologies, model: ICP-MS 7500cx )
- Neubauer-improved hemocytometer (Marienfeld-Superior, catalog number: 0650030 )
Procedure
- Sampling and washing (Figure 1)
Notes: - Keep samples on ice to slow down the metabolic activity.
- The washing steps are time sensitive since they stop the experiment by removing all external Mn. Removing all external Mn conserves the sample in the state of the timepoint it was taken and the elemental composition of the sample cannot change anymore.
- Sample volumes and final elution volumes are accounted for in the calculation under data analysis. All volumes in this protocol are therefore approximate volumes, as long as they are weighted accurately. All transfers can be done by pouring. Pipetting is not necessary and may even add contaminants.
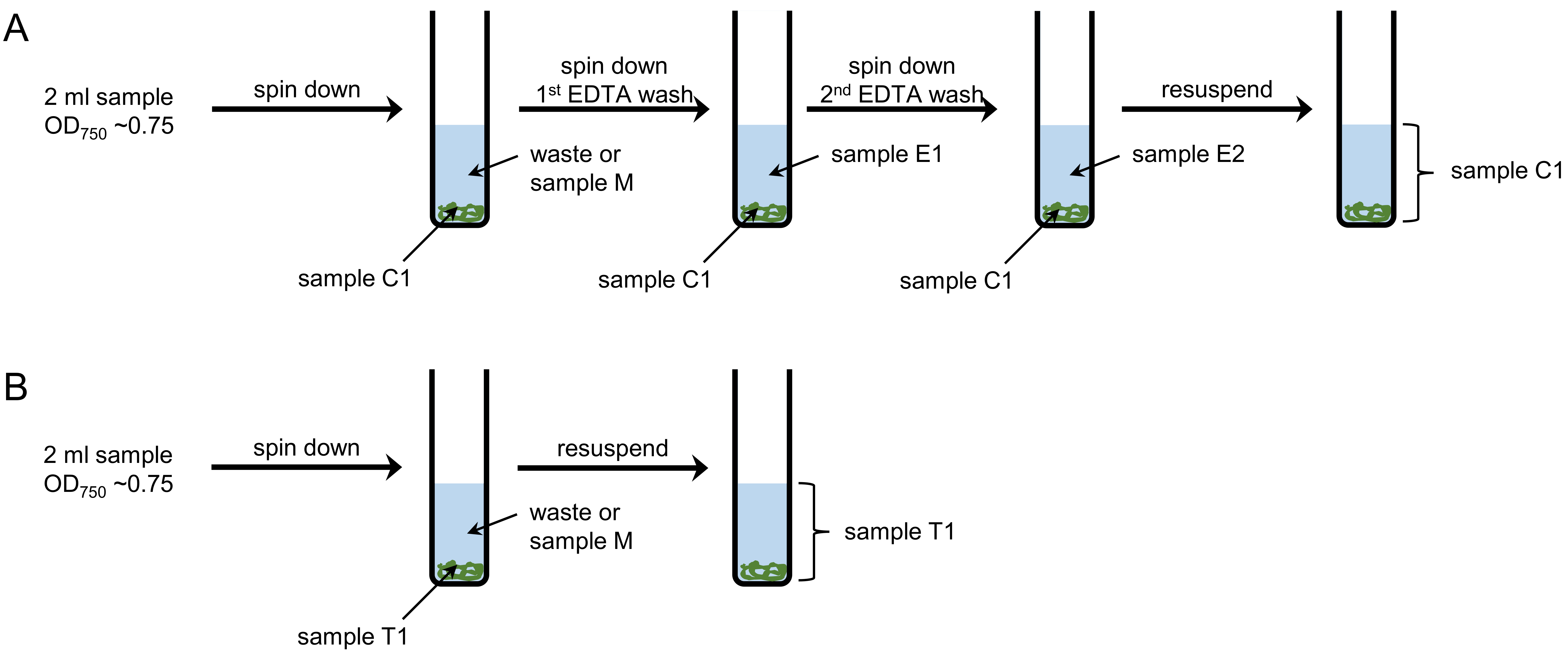
Figure 1. Workflow for sampling and washing. A. Intracellular (sample C) and periplasmic (samples E1 and E2) Mn pools are separated by EDTA washing steps. B. To measure the total cellular Mn content (sample T), cells are separated from the medium and resuspended in Milli-Q water. - For each sample, prepare four 15 ml test tubes, labeled with sample number and T1 (total cell), E1 (first EDTA wash), E2 (second EDTA wash) and C1 (intracellular Mn content), respectively.
- Weigh and note the weight of each test tube, including cap.
- Take 2 ml samples (OD750 ~0.75, Note 3) and transfer them into tubes T1 and C1 (Note 4).
- Remove 30 µl from tube T1 for cell counting.
- Weigh and note the weight of tubes T1 and C1.
Note: Subtracting the weight of the empty tube from that of the tube with sample gives you the weight of the sample. - Centrifuge tubes T1 and C1 at 3,000 x g and 4 °C for 5 min.
- Discard the supernatant.
Note: The supernatant can be analyzed as sample M (medium) if wanted. If so, weigh the sample before filtering it through a 0.2 µm syringe filter to get rid of remaining cells. The growth of remaining cells would change the Mn composition of the medium and require sample digestion. After filtering, the sample does not need further processing. - Resuspend samples T1 by vortexing in approximately 4 ml of Milli-Q water. Store the samples at room temperature until sample digestion.
- Add 4 ml EDTA wash buffer to samples C and resuspend by vortexing. Repeat step 1f.
- Transfer the EDTA supernatant to the test tube labeled E1. Weigh and note the weight. Store tube E1 at room temperature. The sample does not need further processing (Note 5).
Note: Most ICP-MS facilities do not accept undigested or unfiltered samples. Therefore, it may be necessary to filter the EDTA washes as described above. However, potentially remaining cells cannot grow, since they are in EDTA wash buffer and not medium. - Add 4 ml EDTA wash buffer to samples C and resuspend by vortexing. Repeat step 1f.
- Transfer the EDTA supernatant to the test tube labeled E2. Weigh and note the weight. Store tube E2 at room temperature. The sample does not need further processing.
- Add 4 ml Milli-Q water and resuspend by vortexing.
- Remove 30 µl from the tube for cell counting.
- Sample digestion
Note: From here on, it is essential to carry out all steps in clean room conditions.- For each sample, prepare another set of two 15 ml test tubes (label with sample number and C2 or T2).
- Weigh out 4 ml of Milli-Q water to the test tubes C2 and T2
- Add 2 ml of nitric acid to each of samples T1 and C1.
- Gently shake or invert for 5 min.
- Preheat a hot plate in a chemical hood to 200 °C (Note 6).
- Place each sample (T1 and C1 only) into Teflon cups suitable for the hot plate and place the samples on the hot plate.
- Let the samples evaporate until minimal droplet remains; remove from the hot plate before sample evaporates completely (Note 7).
- Add all water from corresponding test tube to the Teflon cup, re-suspend the sample carefully and return the sample to the same test tube (e.g., water from tube T2 to the evaporated sample T and back to tube T2).
- For each sample, prepare another set of two 15 ml test tubes (label with sample number and C2 or T2).
- Sample measurement (clean room not required)
- Transfer 2 ml from each sample (T2, C2, E1, and E2) to appropriate sample tubes for the ICP-MS sample handler.
- Store the remaining 2 ml of each sample for potential extra measurements.
- Analyze the samples according to the instructions of your ICP-MS facility.
- Transfer 2 ml from each sample (T2, C2, E1, and E2) to appropriate sample tubes for the ICP-MS sample handler.
- Cell counting
- Perform cell counting on the samples from sampling and washing (steps 1d and 1n) according to the instructions manual of your hemocytometer.
- Perform cell counting on the samples from sampling and washing (steps 1d and 1n) according to the instructions manual of your hemocytometer.
Data analysis
The periplasmic Mn concentration is the sum of the Mn in samples E1 and E2. Sample C2 is the intracellular concentration of Mn, namely cytosol and thylakoid system. Apply equation 1 to calculate the periplasmic Mn concentration using samples E1, E2 as well as volume and cell count from C1. Similarly, use equation 2 to calculate the intracellular Mn concentration in [atoms cell-1] using samples C1 and C2. 
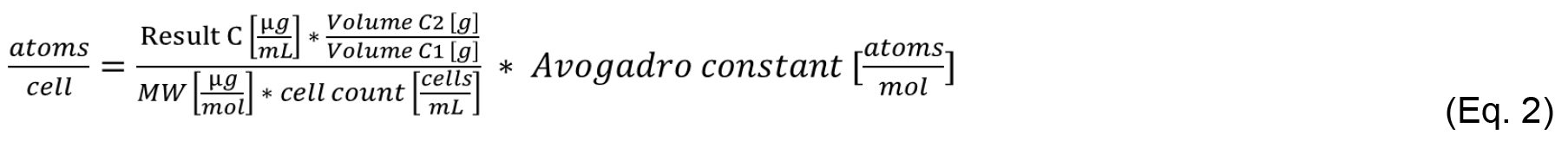
ICP-MS results usually come in parts per million (ppm), which is equivalent to [µg ml-1] or parts per billion (ppb), which is equivalent to [µg L-1]. Make sure to convert the input data into the correct units. Please note that the molecular weight (MW) in equations 1 and 2 is in [µg mol-1].
Instead of cell counting, you can use OD750 or chlorophyll content for normalization as well. However, we recommend cell counting. As a control, the sum of the Mn in samples C2, E1 and E2 should equal the Mn concentration in sample T2. You can calculate sample T2 in the same way as sample C2.
Figure 2 shows an example of the calculations. The numbers used for this calculation were obtained from a WT sample grown at our standard conditions (Note 1).
Figure 2. Example calculation of Mn concentration. Make sure the input data is in the correct units. In our case, for example, the ICP-MS results were in [µg L-1]. Please note that the molecular weight (MW) of Mn is in [µg mol-1]. In our experiments, the difference between E1 + E2 + C2 and T2 was in the range of 0.1-10 x 103 [atoms cell-1].
Notes
- Mutant lines are based on and compared to a Japanese WT of Synechocystis sp. PCC 6803 obtained from Martin Hagemann (University of Rostock, Germany). Our standard growth conditions are 30 °C, 100 rpm shaking, and 100 µmol photons m-2 sec-1.
- This is an elemental analysis that does not differentiate between Mn incorporated into proteins, complexed Mn and free Mn.
- The amount of original sample to be analyzed depends on the cell density of the sample. Dense samples require less volume and dilute samples require more. There is no need to adjust the other volumes. In our experiments, we took 2 ml samples with an OD750 ~0.75.
- There is no need to use a pipette following this protocol. This minimizes the contamination of the sample. All transfers from one tube to another should be done by pouring.
- In principle, it is possible to combine samples E1 and E2 and measure them together. If doing so, the sample volume in the calculation needs then to be changed accordingly. However, depending on the physiological conditions of the experiment, the amount of Mn found in these samples can be close to the detection limit already in sample E1. Therefore, we prefer to not further dilute sample E1 with sample E2.
- The temperature of the hot plate can be set as low as 180 °C if handling a large number of samples. Doing this will slow down the process allowing for better tracking of all samples on the hot plate to avoid burning.
- The minimal possible droplet size mostly depends on the experience of the person conducting the experiment. Importantly, the droplet size gets included in the sample weight before the measurements meaning larger droplets do not affect the measurements. This step is solely to protect the ICP-MS pump by reducing the amount of nitric acid in the sample. If samples do evaporate completely, add 1 ml of nitric acid and mix well. Continue evaporation carefully.
- All samples can be stored at room temperature until digestion, as soon as the experiment has stopped by the washing steppes. All possible contaminations will be eliminated in the digestion process but cannot change the elemental composition of the sample. We are not aware of any specific shelf life of the samples before digestion, however, the longest time we stored samples before digestion was three weeks. After digestion, we recommend freezing the samples to prevent microbial growth which may affect the ICP-MS machine rather than the sample composition.
Recipes
- EDTA wash buffer
20 mM HEPES-KOH, pH 7.5
5 mM EDTA
Note: EDTA wash buffer can be stored at room temperature for several months.
Acknowledgments
This research was supported by the German Science Foundation (EI 945/3-1 to M.E.) and the Israeli Science Foundation (2733/16 to N.K.). The authors declare that there are no conflicts of interest.
References
- Brandenburg, F., Schoffman, H., Kurz, S., Kramer, U., Keren, N., Weber, A. P. and Eisenhut, M. (2017). The Synechocystis manganese exporter Mnx is essential for manganese homeostasis in cyanobacteria. Plant Physiol 173(3): 1798-1810.
- Keren, N., Kidd, M. J., Penner-Hahn, J. E. and Pakrasi, H. B. (2002). A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry 41(50): 15085-15092.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Brandenburg, F., Schoffman, H., Keren, N. and Eisenhut, M. (2017). Determination of Mn Concentrations in Synechocystis sp. PCC6803 Using ICP-MS. Bio-protocol 7(23): e2623. DOI: 10.21769/BioProtoc.2623.
- Brandenburg, F., Schoffman, H., Kurz, S., Kramer, U., Keren, N., Weber, A. P. and Eisenhut, M. (2017). The Synechocystis manganese exporter Mnx is essential for manganese homeostasis in cyanobacteria. Plant Physiol 173(3): 1798-1810.
Category
Microbiology > Microbial biochemistry > Other compound
Microbiology > Microbial metabolism > Nutrient transport
Biochemistry > Other compound > Elements
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









