- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Design and Direct Assembly of Synthesized Uracil-containing Non-clonal DNA Fragments into Vectors by USERTM Cloning
Published: Vol 7, Iss 22, Nov 20, 2017 DOI: 10.21769/BioProtoc.2615 Views: 10331
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Rapid and Cost-Effective Pipeline to Identify and Capture BGCs From Bacterial Draft Genomes
Marco A. Campos-Magaña [...] Luis Garcia-Morales
Dec 20, 2025 766 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 543 Views
Abstract
This protocol describes how to order and directly assemble uracil-containing non-clonal DNA fragments by uracil excision based cloning (USER cloning). The protocol was generated with the goal of making synthesized non-clonal DNA fragments directly compatible with USERTM cloning. The protocol is highly efficient and would be compatible with uracil-containing non-clonal DNA fragments obtained from any synthesizing company. The protocol drastically reduces time and handling between receiving the synthesized DNA fragments and transforming with vector and DNA fragment(s).
Keywords: USERTM cloningBackground
For synthesized DNA, non-clonal linear DNA fragments (NCDF) have emerged as a cheaper and faster alternative to clonal fragments that are delivered sequenced and in a circular vector. NCDFs can be regarded as an IKEA solution to DNA synthesis where it is the customers that assemble their DNA fragments into a vector of choice and subsequently must verify the sequence of the final construct. In this protocol, we obtained uracil-containing NCDFs from Thermo Fisher Scientific Geneart, whose NCDFs are termed DNA strings. The uracil-containing NCDFs will here be named uNCDFs. In uNCDFs, uracils are inserted at designated positions during synthesis. We were able to clone uNCDFs directly into USER cloning compatible vectors by simply re-suspending the fragments in water, incubating them with linearized USER vector and USER enzyme (as described below) followed by transformation of E. coli. The procedure requires minimal handling, lasts less than 1 h from receipt to transformation and requires very small amounts of DNA. We have tested uNCDFs for almost a year in our laboratory and found them to perform consistently well. We recently cloned 13 transporter genes into a USER compatible Xenopus oocyte expression vector. Each gene fragment was obtained as a uNCDF (~2 kb in size). We tested a single colony for each cloning and found that 12 of the genes were inserted correctly into the pNB1u destination vector. For the last clone we found a single mutation which was correct in the second colony that we tested for that fragment (Jorgensen et al., 2017). The pNB1u vector is approximately 2.5 kb in size. In another project, we cloned a larger batch of uNCDFs composed of 64 fragments 1-2.4 kb in size into a variety of USER compatible vectors including the large (~10 kb) USER compatible pCambia based vectors (Nour-Eldin et al., 2006). Of these, 55 were correct after testing a single colony, 62 were correct after sequencing a second colony, while the remaining two were found in a correct form in the third colony (unpublished). We also used the protocol to seamlessly fuse three fragments together and insert them into a destination vector by USER fusion (Geu-Flores et al., 2007). Thus, we experienced high efficiency when cloning uNCDFs which was comparable to the efficiency of cloning uracil-containing DNA fragments generated via PCR. Moreover the uNCDFs were shown to work well for both small and large USER compatible vectors.
The innovation in this protocol is the ability to design and order synthesized DNA fragment containing uracils at appropriate locations. To appreciate the value of this advance and ensure an understanding of this cloning technique for researchers who have not used USER cloning previously, a brief introduction to USER cloning is given below.
The principle behind the efficiency of USER cloning lies in the ability to generate long, complementary overhangs. These overhangs can anneal to form stable hybridization products that can be used to transform E. coli without prior ligation. Most importantly, their generation is not dependent on the introduction of restriction sites. Hitherto, the generation of long single stranded overhangs has proceeded in two steps. First, a PCR reaction was performed on a DNA fragment of interest using primers containing a short 8-16 nt upstream extension that is preceded by a single deoxyuridine residue. The resulting PCR products are treated briefly with a commercial mix of uracil DNA glycosylase and DNA glycosylaselyase Endo VIII. These enzymes, are included in the USERTM enzyme mix, and remove the two single deoxyuridine residues and enable the dissociation of the short, single-stranded fragments lying upstream from the cleavage sites. For the generation of overhangs in a USER compatible destination vector, a short cassette is inserted into it so that digestion with a restriction- and a nicking enzyme creates the desired long overhangs. The fact that long, custom-made overhangs can be generated on PCR products can be exploited to generate a series of PCR products with complementary overhangs. This enables the generation of a hybridization product consisting of a vector and multiple PCR products, which can be fused into a compatible vector as easily as a single PCR product. For more detailed information on the cloning method, readers are referred to the following references on how to perform USER cloning and USER fusion (Nour-Eldin et al., 2006 and 2010; Geu-Flores et al., 2007). Additionally, bioinformatic tools have been developed to aid design of primers and strategy of USER cloning and fusion including the (Automated DNA Modifications with USER cloning) AMUSER web server tool for automated primer design (Genee et al., 2015) and an interactive lab simulation that teaches the principles of USER cloning (https://www.labster.com/simulations/user/). Finally, overlap design for USER fusion has recently been optimized (Cavaleiro et al., 2015).
Introducing uracils into DNA fragments was hitherto performed via PCR using uracil-containing primers. The PCR was performed after receiving the NCDF and required designing and ordering of appropriate uracil-containing primers. Incorporating uracils in DNA fragments during synthesis omits the need to design and order uracil-containing primers and also omits the need to perform a PCR reaction, this drastically reduces time and handling between receiving the synthesized DNA fragments and transforming with vector and DNA fragment(s).
Materials and Reagents
- For ordering uracil-containing NCDFs
- Sequence of DNA to be synthesized (optimized for desired organism)
Note: Depending on the fidelity of synthesis, any size of fragment is in principle suitable for synthesis. Until now the maximum size of NCDFs we have been able to order is 3 kb (varies depending on the company) and the fidelity in terms of how many colonies we had to sequence to find a correct clone has generally been 1-2. - Sequence of USER tails to be added for insertion into USER vector (for single gene insertion)
- Sequence of USER overlaps for seamless USER fusion
Note: For insertion into USER compatible vectors, the USER tails are given by the USER cassette that has been inserted. Please consult the relevant publications. For USER fusion we typically use USER overlaps between 7-16 bp in length. - Sequence of DNA to be synthesized (optimized for desired organism)
- For linerarizing USER compatible vector
- 1.5 ml centrifuge tubes (Eppendorf tubes)
- USER compatible vector (Nour-Eldin et al., 2006 and 2010; Geu-Flores et al., 2007)
- PacI restriction enzyme (New England Biolabs, catalog number: R0547S )
- Nt.BbvCI nicking enzyme (New England Biolabs, catalog number: R0632S )
- Cutsmart buffer (included when ordering enzymes above)
- H2O
- PCR purification kit of choice (e.g., QIAquick PCR purification kit) (QIAGEN, catalog number: 28104 )
Note: For part B, follow protocol published previously (Nour-Eldin et al., 2010). - 1.5 ml centrifuge tubes (Eppendorf tubes)
- For cloning uNCDFs into linearized USER compatible vector
- 1.5 ml centrifuge tubes (Eppendorf tubes)
- CaCl2 competent E. coli cells (DH5α or NEB10β or other common cloning strains)
Note: Do not perform transformation by electroshocking as the shock will cause the vector and uNCDF to dissociate. - uNCDFs from synthesis company (in this case from Thermo Fisher Scientific Geneart)
Note: Despite their success, uNCDFs are not yet offered officially by Geneart. Please contact e.g., Anja Martinez at Thermofisher Geneart (anja.martinez@thermofisher.com) or us for ordering questions. - H2O
- Linearized USER compatible vector
Note: A long list of vectors has been made USER cloning compatible for almost all organisms. Search within these three papers (Nour-Eldin et al., 2006 and 2010; Geu-Flores et al., 2007) and the many papers citing them. Please note, we are currently in the process of gathering USER vectors from groups from all over the world in order to deposit them at Addgene for easy accessibility. - USER enzyme (New England Biolabs, catalog number: M5505S )
- 5x or 10x PCR buffer (any kind)
- 1.5 ml centrifuge tubes (Eppendorf tubes)
Equipment
- 37 °C heat block
- 42 °C heat block/water bath
Procedure
- Design and order uracil-containing NCDFs for single fragment USER cloning
For cloning single uNCDFs into USER compatible vectors that have been generated by our lab, our generic 5’ and 3’ user tails are added to termini of the fragment to be inserted (5’ USER tail: 5’-GGCTTAAU, 3’ USER tail: 5’-GGTTTAAU) (for inserting into other USER compatible vectors please use the corresponding USER tails). When ordering synthesized DNA we typically codon optimize the sequence for the organism wherein the gene will be expressed. In an example below, we show how we ordered the gene AT1G15210 as a uNCDF from Thermo Fisher Scientific Geneart. As instructed by Geneart, we provided them with a word file containing sequences to be synthesized in FASTA format. At the beginning of the word document, we included general instructions to the synthesis company, which are given in italics below
Dear synthesis company:
Green marks a T in the top strand that must be replaced by a U. Turquoise marks an A, which is complementary to a T on the opposite strand that must be replaced by a U (i.e., it is the T on the opposite strand that must be replaced by a uracil). Yellow marks sequence that may not be altered. Non-marked sequence represents coding sequence, which may be changed to overcome synthesis challenges. The gene will be expressed in Xenopus laevis oocytes. Please use the appropriate codon preference table for codon optimization and alternations.
Note: Punctuation marks approx. 2 kb of sequence that was included in the order but which has been omitted here for brevity. In this example, yellow sequence represents our standard 5’ and 3’ USER tails. - Cloning single uNCDFs into linearized USER compatible vector
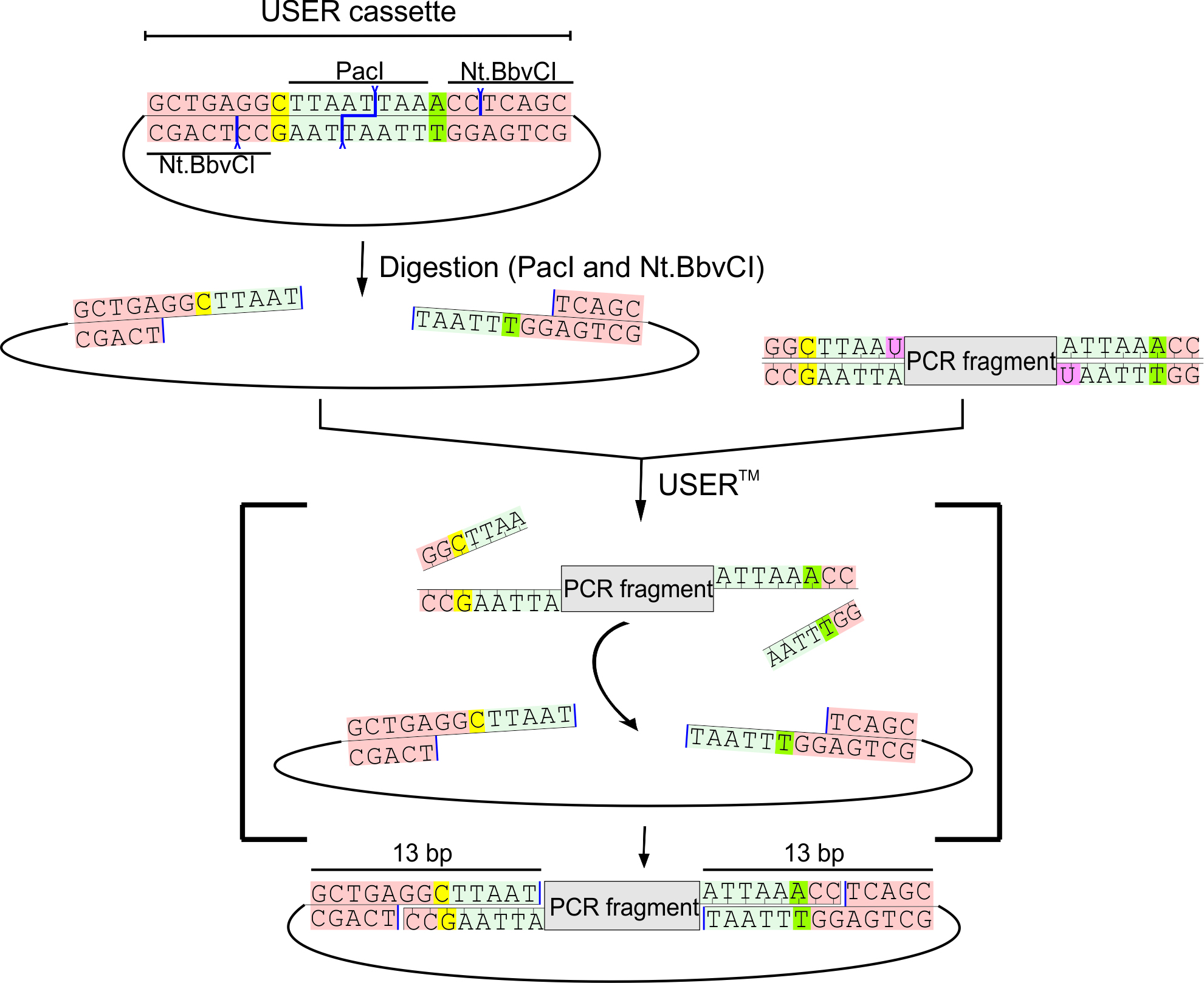
Figure 1. Overview of the USER cloning technique. A USER cassette in a USER-compatible vector (upper left corner) with a restriction site (PacI, light blue) in the middle, one variable nt surrounding it (different for each side, yellow and green), and oppositely oriented nicking sites (Nt.BbvCI, tan). The USER vector is digested with PacI and Nt.BbvCI to generate 8 nt overhangs. A DNA fragment (upper right corner) with uracils at appropriate positions can be generated by PCR as described previously or via synthesis as a uNCDF as described here. Fragment and vector are mixed with USERTM enzyme mix (excising deoxyuridines, pink) and the digested USER-compatible vector. Following brief incubation, the hybridized product can be used to transform E. coli without prior ligation. This figure has been reproduced with permission from (Nour-Eldin et al., 2006).- Each fragment is to be surrounded by USER tails that enable insertion into a USER compatible vector. In this example, we use our X. laevis expression vector pNB1u (Nour-Eldin et al., 2006). uNCDFs are mixed directly with the digested pNB1u vector without prior PCR amplification (Figure1).
- Dilute each uNCDF to 100 ng/μl in H2O (or TE buffer pH 8).
- The USER-compatible pNB1u X. laevis oocyte expression vector is digested with PacI/Nt.BbvCI overnight, PCR purified and diluted to a concentration of ~50 ng/μl (as previously described [Nour-Eldin et al., 2006 and 2010]).
Note: If a USER compatible vector is not available, it is possible to generate them via PCR. In that case, the vector backbone is treated as a DNA fragment to be assembled with DNA fragment of interest via USER fusion (please see below). - For the USER reaction, mix 100 ng uNCDF with 50 ng digested pNB1u, 1 U USER enzyme (NEB), 2 µl 5x PCR reaction buffer and 5 µl H2O.
- Incubate the reaction at 37 °C for 25 min, and then for 25 min at room temperature.
- Transform 50 µl chemically competent E. coli cells with the reaction mixture by heat shock (5 min on ice, 30-45 sec at 42 °C and 5 min on ice).
- Plate the transformation mixture on LB-plates containing appropriate antibiotic selection (for pNB1u we use carbenicillin or ampicilin–containing LB plates).
- Select three colonies from the plates and grow overnight. Extract plasmids and analyze the extracted plasmids by gel-electrophoresis. Sequence the plasmids with an insert.
- Dilute each uNCDF to 100 ng/μl in H2O (or TE buffer pH 8).
- Each fragment is to be surrounded by USER tails that enable insertion into a USER compatible vector. In this example, we use our X. laevis expression vector pNB1u (Nour-Eldin et al., 2006). uNCDFs are mixed directly with the digested pNB1u vector without prior PCR amplification (Figure1).
- Design and order uracil-containing NCDFs for USER fusion of multiple fragments
For fusion of multiple uNCDFs into USER compatible vectors overlap regions have to be selected and included in adjacent fragments. Overlap regions are selected as previously described by finding a T on the bottom strand and a T on the top strand 7-15 bases downstream of the first T. The pair of selected Ts should be within 20-30 bases distance to the junction site (Figure 2) (Geu-Flores et al., 2007; Nour-Eldin et al., 2010). These Ts will be the ones replaced by a U during synthesis. For insertion into the USER compatible vector, our generic 5’ and 3’ user tails are added to the terminal fragments. When ordering synthesized DNA we typically codon optimize the sequence for the organism wherein the gene will be expressed. In an example below, we show how we designed and ordered three fragments to be fused and inserted into a USER compatible vector by USER cloning. The full-length sequence (~7,200 bp) was split into three fragments (FAS1_A, FAS1_B and FAS1_C). The sequence of their termini is given below. In each fragment, the punctuation in the middle denotes approximately 2 kb of sequence, which was included in the order but has been omitted here for brevity. Each fragment was designed to include overlap regions to the adjacent fragment close to the junction sites. Please see below and see (Geu-Flores et al., 2007; Nour-Eldin et al., 2010) for more detailed information on how to design USER fusion overlap regions. Alternatively, the Amuser web server tool can deisgn overlap regions for any USER fusion approach (Genee et al., 2015). As instructed by Geneart we provided them with a Word file containing sequences to be synthesized in FASTA format. At the beginning of the Word document, we included general instructions to the synthesis company, which are given in italics below.
Dear synthesis company:
Green marks a T in the top strand that must be replaced by a U. Turquoise marks an A, which is complementary to a T on the opposite strand that must be replaced by a U (i.e., it is the T on the opposite strand that must be replaced by a uracil). No alterations are allowed in any color shaded sequence. Non-marked sequence represents coding sequence, which may be changed to overcome synthesis challenges. The genes will be expressed in Xenopus laevis oocytes. Please use the appropriate codon preference table for codon optimization and alternations.
Note: Only the sense strand is included in the ordering process but the result is a uracil-containing double stranded fragment. No alterations are allowed in any colored region during the codon optimization process. Yellow marks our standard 5’ USER tail. Green marks the T, which must be synthesized as U. Turquoise marks the A, whose complimentary T must be synthesized as U. Grey marks the overlap which will be exposed as a single stranded fragment upon USER treatment and which will hybridize to the complimentary single stranded overhang on the adjacent fragment (in this case FAS1_B). Once synthesized, the double stranded FAS1_A uNCDF will look as follows:
Upon treatment with USER enzyme the double stranded FAS1_A uNCDF will loose sequences lying upstream of the uracils and look as follows:
As for FAS1_A only the sense strand is included in the ordering process. Green marks the T, which must be synthesized as U. Turquoise marks the A, whose complimentary T must be synthesized as U. Grey marks the overlaps which will be exposed as single stranded overhangs upon USER treatment and which will hybridize to the complimentary single stranded overhangs on the adjacent fragments (in this case the left ‘grey’ will hybridize to the overhang generated on FAS1_A, whereas the right ‘grey’ will hybridize to the overhang generated on FAS1_C). Once synthesized, the double stranded FAS1_B uNCDF will look as follows:
Upon treatment with USER enzyme the double stranded FAS1_B uNCDF will loose sequences lying upstream of the uracils and look as follows:
As for FAS1_A only the sense strand is included in the ordering process. Green marks the T, which must be synthesized as U. Turquoise marks the A, whose complimentary T must be synthesized as U. Grey marks the overlap, which will be exposed as single stranded overhang upon USER treatment and which will hybridize to the complimentary single stranded overhangs on the adjacent fragment (in this case the rightmost ‘grey’ overlap region on FAS1_B). Once synthesized, the double stranded FAS1_B uNCDF will look as follows:
Upon treatment with USER enzyme the double stranded FAS1_C uNCDF will loose sequences lying upstream of the uracils and look as follows:
- Fusion and clone of multiple uNCDFs into linearized USER compatible vector
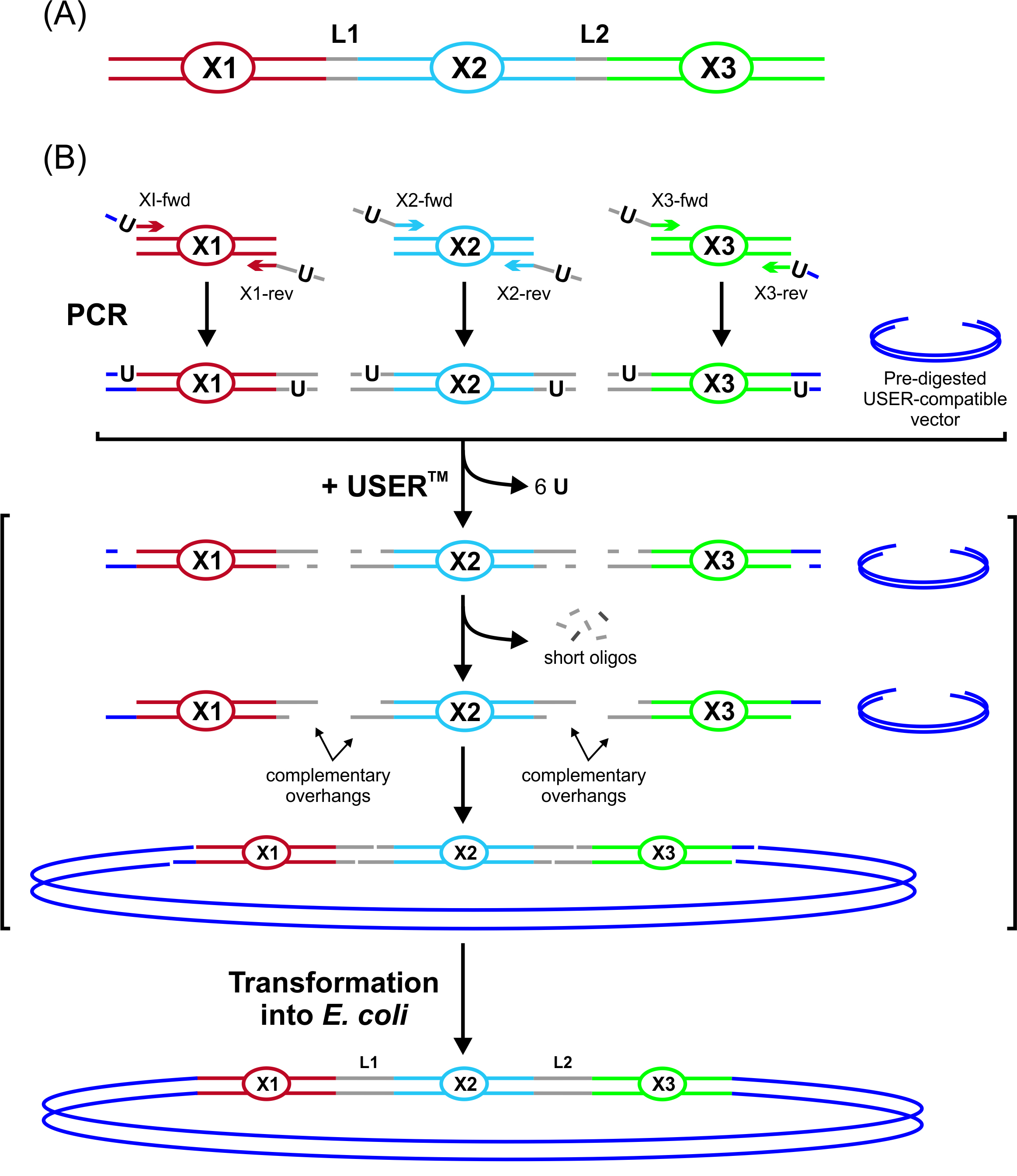
Figure 2. Overview of the USER fusion technique. Fragments X1, X2, and X3 are to be fused together. Overlap regions are marked in grey. Uracils can be inserted via PCR or as described here during synthesis. The DNA fragments are mixed with a pre-digested USER-compatible vector and treated with the deoxyuridine-excising USERTM enzyme mix. This generated 3’ overhangs that complement each other (indicated by arrows), while the outermost ones complemented the overhangs of the pre-digested vector. This design enables the formation of a stable circular hybridization product that can be transformed directly into E. coli without prior ligation. This figure has been reproduced with permission from (Geu-Flores et al., 2007).
For fusing and cloning multiple uNCDFs into USER compatible vectors that have been generated by our lab, our generic 8 bp long 5’ and 3’ user tails are added to termini of the terminal fragments. At junction sites uracil-containing overlaps were included to allow seamless fusion upon mixing. When possible we strive to choose fusion overlaps of different lengths and composition to minimize mis-hybridization between fragments. In this example, we fuse and clone three fragments into our X. laevis expression vector pNB1u (Nour-Eldin et al., 2006). Each fragment contained a uracil at the appropriate location in each USER tail. The uracil was incorporated during synthesis. Thus, uNCDFs are mixed in equimolar ratios directly with the digested pNB1u vector without prior PCR amplification with uracil-containing primers.- Dilute each uNCDFs to 50 ng/µl in H2O.
- The USER-compatible pNB1u X. laevis oocyte expression vector is digested with PacI/Nt.BbvCI overnight, PCR purified and diluted to a concentration of ~50 ng/µl (as previously described (Nour-Eldin et al., 2006 and 2010)).
- For the USER reaction, mix 100 ng of each uNCDFs with 50 ng digested pNB1u, 1 U USER enzyme (NEB), 2 µl 5x PCR reaction buffer and H2O to bring the total volume to 10 µl.
- Incubate the reaction at 37 °C for 25 min, and then for 25 min at room temperature.
- Transform 50 µl chemically competent E. coli cells (for example NEB10β by heat shock) with the reaction mixture (5 min on ice, 30-45 sec at 42 °C and 5 min on ice).
Note: Typically the volume of added USER reaction mixture should not exceed 10% of the competent E. coli volume (i.e., max 5 µl reaction mixture to 50 µl E. coli volume, 10 µl reaction mixture to 100 µl E. coli volume etc. Exceeding this ration can reduce transformation efficiency. - Plate the transformation mixture on LB-plates containing appropriate antibiotic selection (for pNB1u we used carbenicillin–containing LB plates).
- Select eight colonies from the plates and grow overnight. Extract plasmids and analyze the extracted plasmids by gel-electrophoresis. Sequence the plasmids with an insert.
- Dilute each uNCDFs to 50 ng/µl in H2O.
Notes
If insertion does not succeed in the first trial, we encourage users–in addition to the USER cloning–to clone the uNCDFs into a blunt-ended cloning vector (such as pJET) for safekeeping and future work.
Acknowledgments
MEJ is supported by a grant from the Danish Council for Independent Research: DFF–6108-00122. DX, AP, CW, DV, SKL and HHN were funded by DNRF99 grant from the Danish National Research Foundation. ZNB was funded by Innovationfund Denmark J.nr.: 76-2014-3 and NW was funded by Human Frontier Science Program RGY0075/2015. The authors declare no conflicts of or competing interest.
References
- Cavaleiro, A. M., Kim, S. H., Seppala, S., Nielsen, M. T. and Norholm, M. H. (2015). Accurate DNA assembly and genome engineering with optimized uracil excision cloning. ACS Synth Biol 4(9): 1042-1046.
- Genee, H. J., Bonde, M. T., Bagger, F. O., Jespersen, J. B., Sommer, M. O., Wernersson, R. and Olsen, L. R. (2015). Software-supported USER cloning strategies for site-directed mutagenesis and DNA assembly. ACS Synth Biol 4(3): 342-349.
- Geu-Flores, F., Nour-Eldin, H. H., Nielsen, M. T. and Halkier, B. A. (2007). USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res 35(7): e55.
- Jorgensen, M. E., Xu, D., Crocoll, C., Ramirez, D., Motawia, M. S., Olsen, C. E., Nour-Eldin, H. H. and Halkier, B. A. (2017). Origin and evolution of transporter substrate specificity within the NPF family. Elife 6: e19466.
- Nour-Eldin, H. H., Geu-Flores, F. and Halkier, B. A. (2010). USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. Methods Mol Biol 643: 185-200.
- Nour-Eldin, H. H., Hansen, B. G., Norholm, M. H., Jensen, J. K. and Halkier, B. A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34(18): e122.
Article Information
Copyright
Jørgensen et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Jørgensen, M. E., Wulff, N., Nafisi, M., Xu, D., Wang, C., Lambertz, S. K., Belew, Z. M. and Nour-Eldin, H. H. (2017). Design and Direct Assembly of Synthesized Uracil-containing Non-clonal DNA Fragments into Vectors by USERTM Cloning. Bio-protocol 7(22): e2615. DOI: 10.21769/BioProtoc.2615.
- Jorgensen, M. E., Xu, D., Crocoll, C., Ramirez, D., Motawia, M. S., Olsen, C. E., Nour-Eldin, H. H. and Halkier, B. A. (2017). Origin and evolution of transporter substrate specificity within the NPF family. Elife 6.
Category
Plant Science > Plant molecular biology > DNA
Molecular Biology > DNA > DNA cloning
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










