- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cytosolic and Nuclear Delivery of CRISPR/Cas9-ribonucleoprotein for Gene Editing Using Arginine Functionalized Gold Nanoparticles
Published: Vol 7, Iss 20, Oct 20, 2017 DOI: 10.21769/BioProtoc.2586 Views: 13195
Reviewed by: David CisnerosAndrea PuharAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Multiplex Genome Editing of Human Pluripotent Stem Cells Using Cpf1
Haiting Ma
Nov 20, 2024 2650 Views

Reprogramming White Fat Cells for Adipose Manipulation Transplantation (AMT) Therapy
Kelly An [...] Nadav Ahituv
Aug 5, 2025 2240 Views

Generation of Insulin-Producing Alpha TC1-6 Cells Using EpiCRISPR System for Targeted DNA Methylation
Marija B. Đorđević [...] Melita S. Vidaković
Oct 20, 2025 1232 Views
Abstract
In this protocol, engineered Cas9-ribonucleoprotein (Cas9 protein and sgRNA, together called Cas9-RNP) and gold nanoparticles are used to make nanoassemblies that are employed to deliver Cas9-RNP into cell cytoplasm and nucleus. Cas9 protein is engineered with an N-terminus glutamic acid tag (E-tag or En, where n = the number of glutamic acid in an E-tag and usually n = 15 or 20), C-terminus nuclear localizing signal (NLS), and a C-terminus 6xHis-tag. [Cas9En hereafter]
To use this protocol, the first step is to generate the required materials (gold nanoparticles, recombinant Cas9En, and sgRNA). Laboratory-synthesis of gold nanoparticles can take up to a few weeks, but can be synthesized in large batches that can be used for many years without compromising the quality. Cas9En can be cloned from a regular SpCas9 gene (Addgene plasmid id = 47327), and expressed and purified using standard laboratory procedures which are not a part of this protocol. Similarly, sgRNA can be laboratory-synthesized using in vitro transcription from a template gene (Addgene plasmid id = 51765) or can be purchased from various sources.
Once these materials are ready, it takes about ~30 min to make the Cas9En-RNP complex and 10 min to make the Cas9En-RNP/nanoparticles nanoassemblies, which are immediately used for delivery (Figure 1). Complete delivery (90-95% cytoplasmic and nuclear delivery) is achieved in less than 3 h. Follow-up editing experiments require additional time based on users’ need.
Synthesis of arginine functionalized gold nanoparticles (ArgNPs) (Yang et al., 2011), expression of recombinant Cas9En, and in vitro synthesis of sgRNA is reported elsewhere (Mout et al., 2017). We report here only the generation of the delivery vehicle i.e., the fabrication of Cas9En-RNP/ArgNPs nanoassembly.
Background
Delivery of Cas9-ribonucleoprotein provides an alternative strategy for CRISPR gene delivery, offering a transient way of editing genes. Although a few strategies for Cas9-RNP delivery have been reported, these strategies suffer from endosomal entrapment of both Cas9 protein and sgRNA (Liu et al., 2015). Mechanical methods including membrane deformation (Han et al., 2015), electroporation (Schumann et al., 2015), and the use of hypertonic agents (D’Astolfo et al., 2015) provide direct delivery, however, they require specialized instrumentations and are generally not practical for in vivo therapeutic applications. Our protocol provides an approach for direct cytoplasmic and nuclear delivery of Cas9-RNP that can find applications in both gene editing and genome imaging.
Materials and Reagents
- Round bottom 35 mm confocal dish (MATTEK, catalog number: P35G-0-14-C )
- 24-well plates (Corning, Costar®, catalog number: 3524 )
- Sterile 1.5 ml tubes (Fisher Scientific, catalog number: 05-408-129 )
- Sterile pipette tips
- Cell lines (i.e., HeLa)
- Stock solution of ArgNP gold nanoparticles (~50 μM in water), freshly purified Cas9En protein (~10-20 μM), and sgRNA (~150 μM)
- 1x phosphate-buffered saline (PBS) (GE Healthcare, HyCloneTM, catalog number: SH30028.02 )
- Plain DMEM media (No serum and antibiotics, appropriate media for cell culture, i.e., HeLa cells) (Thermo Fisher Scientific, GibcoTM, catalog number: 10567014 )
- Alexa Fluor 488 NHS Ester (Thermo Fisher Scientific, InvitrogenTM, catalog number: A20000 )
- AmpliScribe T7-Flash- Transcription Kit (Epicentre, catalog numbers: ASF3257 and ASF3507 )
Equipment
- Pipettes
- Cell culture incubator at 5% CO2 and 37 °C
- Fluorescence microscope (any confocal microscope)
Procedure
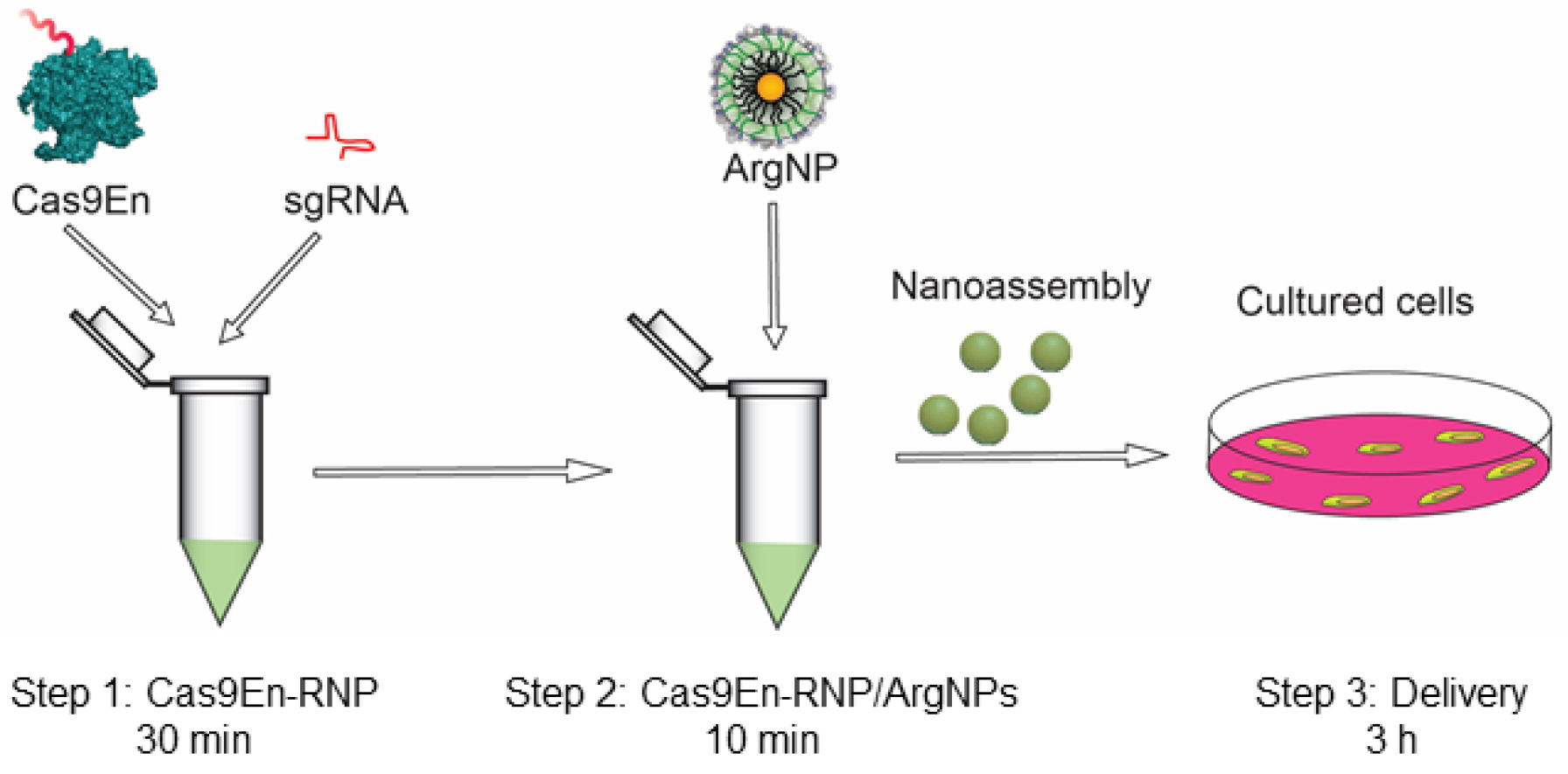
Figure 1. Schematic overview of the protocol. Step 1: formation of Cas9En-sgRNA complex (takes 30 min); Step 2: formation of Cas9En-RNP/ArgNPs nanoassembly (takes 10 min); and Step 3: Cas9En-RNP delivery (takes 3 h).
Final working nanoassembly concentration is 125 nM of ArgNPs and 62 nM of Cas9En-RNP complex, which is at a 2:1 molar ratio of ArgNPs/Cas9En-RNP. The total volume of the nanoassembly samples required for delivery depends on the kind of cell culture plate used. We generally use 1 ml for round bottom 35 mm confocal dishes, 500 μl for 24-well plates, and 200 μl for 96-well plates, per well. Therefore, the nanoassemblies should be made and scaled up according to users’ need. The following calculation is for one sample in a 24-well plate (i.e., 500 μl total volume). Additionally, the following protocol is for HeLa cells, however, we verified the Cas9En-RNP delivery in other cell lines including mouse macrophage RAW 264.7, human embryonic kidney HEK cells, and human primary mammary epithelial cells.
- First, 24 h before the delivery experiment, seed cells on a 24-well plate at a cell density of 8 x 104-10 x 105 cells/well.
- On the day of delivery, prepare the Cas9En-RNP complex by simply mixing Cas9En and sgRNA at a 1:1 molar ratio in 10-20 μl of 1x PBS buffer. Keep in mind that the final Cas9En-RNP concentration in 500 μl sample should be 62 nM (~5 μg Cas9En, and 1 μg sgRNA).
- Incubate the complex at room temperature for 30 min.
- In the meantime, add 100 μl of 1x PBS into a sterile 1.5 ml tube.
- Add required amount of ArgNPs into the tube containing 1x PBS to make the final concentration of ArgNPs 125 nM for 500 μl sample. Mix by pipetting up and down.
- Now add the premixed Cas9En-RNP complex to the ArgNPs solution in the tube.
- Mix well by pipetting up and down.
- Incubate the sample at room temperature for 10 min. The nanoassemblies will be ready in 10 min and therefore users should prepare the cells for delivery in the meantime.
Note: Don’t incubate for more than 10 min, as this may cause aggregate formation instead of well-defined assemblies.
- Wash the cells with 1x PBS, twice.
- When the nanoassembly sample is ready after 10 min, add ~400 μl of DMEM plain media (or any media of interest) into the nanoassembly solution in the tube. Mix well.
Note: Media with serum should be strictly avoided at this step; media with serum may require further optimization such as the ratio of nanoparticles to Cas9En:sgRNA, and incubation time.
- Add the whole (500 μl) sample into the washed-cells.
- Incubate the cells at an appropriate condition (usually 5% CO2 and 37 °C) for 3 h. Complete Cas9En-RNP delivery should be achieved after 3 h of incubation.
- Wash away the media after 3 h, and replace with fresh desired media (DMEM with 10% serum and 1% antibiotics, and any supplements if needed).
Data analysis
Cytoplasmic delivery efficiency of Cas9En or Cas9En-RNP was determined by confocal microscopy imaging. Even distribution of fluorophore labelled Cas9En in the cytosol and nucleus is considered as effective delivery, whereas any punctate distribution in the cytoplasm is considered as endosomal delivery (Figures 2A and 2B). Briefly, ~400 cells were counted manually to estimate effective cytosolic and nuclear delivery. Any punctate distribution (see Figure 2B) of labelled-Cas9En should be avoided from counting as direct cytoplasmic and nuclear delivery. Please see Mout et al., 2017 for more details.
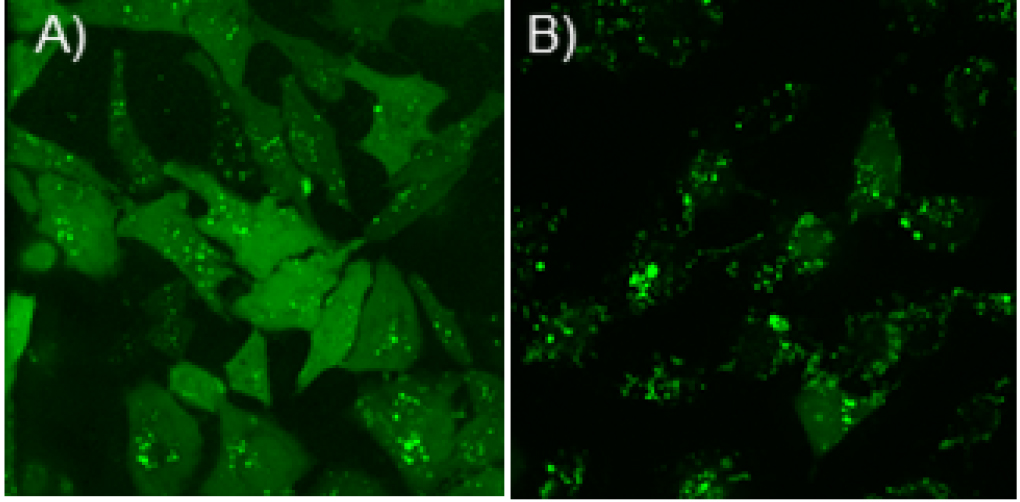
Figure 2. Confocal microscopy images showing examples of efficient cytosolic and nuclear delivery of Cas9E20 (Alexa Fluor 488 labelled) in cultured HeLa cells. A. High efficient cytoplasmic delivery at a 2:1 molar ratio of ArgNP/Cas9E20; B. Low efficient delivery of Cas9E20 that occurs through endocytosis at a 1:1 molar ratio of ArgNP/Cas9E20. Note that, maximum cytoplasmic delivery efficiency can be achieved through screening different molar ratio of ArgNPs/Cas9En or Cas9En-RNP, as noted below.
Notes
- As a point of reference, gene editing verification is performed 48 h after nanoassembly treatment.
- The ratio ArgNPs/Cas9En is the most crucial factor for delivery efficiency. Every batch of ArgNPs can be slightly deferent in terms of surface ligand coverage and therefore users are suggested to test different ratios of ArgNPs/Cas9En to find out maximum delivery. Fluorophore labelled (FITC/Alexa Fluor 488) Cas9En should be used to evaluate delivery efficiency (see manufacturer’s protocol for labelling: Alexa Fluor 488 NHS Ester, Thermo Fisher Scientific).
- Freshly prepared (not more than a week old, and preserved at 4 °C) Cas9En should be used for better activity. Flash-frozen Cas9En at -80 °C with glycerol should be avoided as our preliminary investigation shows that glycerol hampers assembly formation.
Acknowledgments
This research was supported by the NIH (GM077173), NSF (CHE-1307021) and a UMass OTCV grant.
References
- D’Astolfo, D. S., Pagliero, R. J., Pras, A., Karthaus, W. R., Clevers, H., Prasad, V., Lebbink, R. J., Rehmann, H. and Geijsen, N. (2015). Efficient intracellular delivery of native proteins. Cell 161(3): 674-690.
- Han, X., Liu, Z., Jo, M. C., Zhang, K., Li, Y., Zeng, Z., Li, N., Zu, Y. and Qin, L. (2015). CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci Adv 1(7): e1500454.
- Liu, J., Gaj, T., Yang, Y., Wang, N., Shui, S., Kim, S., Kanchiswamy, C. N., Kim, J. S. and Barbas, C. F., 3rd (2015). Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat Protoc 10(11): 1842-1859.
- Mout, R., Ray, M., Tonga, G. Y., Lee, Y. W., Tray, T., Sasaki, K. and Rotello, V. M. (2017). Direct cytoplasmic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 11(3): 2452-2458.
- Schumann, K., Lin, S., Boyer, E., Simeonov, D. R., Subramaniam, M., Gate, R. E., Haliburton, G. E., Ye, C. J., Bluestone, J. A., Doudna, J. A. and Marson, A. (2015). Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A 112(33): 10437-10442.
- Yang, X. C., Samanta, B., Agasti, S. S., Jeong, Y., Zhu, Z. J., Rana, S., Miranda, O. R. and Rotello, V. M. (2011). Drug delivery using nanoparticle-stabilized nanocapsules. Angew Chem Int Ed Engl 50(2): 477-481.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mout, R. and Rotello, V. M. (2017). Cytosolic and Nuclear Delivery of CRISPR/Cas9-ribonucleoprotein for Gene Editing Using Arginine Functionalized Gold Nanoparticles. Bio-protocol 7(20): e2586. DOI: 10.21769/BioProtoc.2586.
Category
Cell Biology > Cell engineering > CRISPR-cas9
Molecular Biology > DNA > DNA modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link