- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
NP-40 Fractionation and Nucleic Acid Extraction in Mammalian Cells
Published: Vol 7, Iss 20, Oct 20, 2017 DOI: 10.21769/BioProtoc.2584 Views: 15300
Reviewed by: Gal HaimovichAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Nuclei from Human Snap-frozen Liver Tissue for Single-nucleus RNA Sequencing
Marcus Alvarez [...] Päivi Pajukanta
Feb 5, 2023 2594 Views

Fast and Sustainable Thermo-osmotic DNA Extraction Protocol for Trans-spectrum Contingency and Field Use
Stavroula Goudoudaki [...] Yiannis Manoussopoulos
Sep 5, 2023 1726 Views

Nuclei Isolation Methods on Frozen Clotted Blood Samples
Melissa Cuevas [...] Nancy Hadley Miller
Jan 20, 2026 306 Views
Abstract
This technique allows for efficient, highly purified cytoplasmic and nuclear-associated compartment fractionation utilizing NP-40 detergent in mammalian cells. The nuclear membrane is not disturbed during the fractionation thus leaving all nuclear and perinuclear associated components in the nuclear fraction. This protocol has been modified from Sambrook and Russell (2001) in order to downscale the amount of cells needed. To determine the efficiency of fractionation, we recommend using qPCR to compare the subcellular compartments that have been purified with equivalent amount of control whole cell extracts.
Keywords: Cell fractionationBackground
To fully obtain an understanding of cellular processes, fractionation of nuclear and cytoplasmic compartments are needed. There are many protocols and even some commercial kits available to help separate the two compartments. However, most require high centrifugation speeds and there is a high discrepancy in the yield and even the methods to verify the amount of contamination in the final products. Our protocol utilizes a small benchtop centrifuge at low speeds to obtain highly pure extractions for the cytoplasmic and a combined nuclear/perinuclear associated compartments as well as the data analysis to verify the percentage of contamination. To date, the cells lines that have been tested are 293 T, HeLa and GHOST cell lines. (Galvis, 2014; Galvis et al., 2014).
Materials and Reagents
- BD 21 G needles (Fisher Scientific, catalog number: 14-823-55)
Manufacturer: BD, catalog number: 305274 . - BD tuberculin syringes (Fisher Scientific, catalog number: 14-823-2F)
Manufacturer: BD, catalog number: 309602 . This product has been discontinued. - TipOne 10 µl pipet tips (USA Scientific, catalog number: 1111-3200 )
- TipOne 1-200 µl natural pipet tips (USA Scientific, catalog number: 1111-1800 )
- TipOne 1,000 µl natural pipet tips (USA Scientific, catalog number: 1111-2020 )
- Costar 6-well cell culture plates (Fisher Scientific, catalog number: 07-200-80)
Manufacturer: Corning, catalog number: 3506 . - 1.5 ml microcentrifuge tubes (Fisher Scientific, catalog number: 14-666-318 )
- Iscove’s modification of DMEM (Mediatech, catalog number: 10-016-CV )
- Tween 40 (CHEM-IMPEX INTERNATIONAL, catalog number: 01513 )
- 1x phosphate buffered saline (PBS) (MP Biomedicals, catalog number: 091860454 )
- 1x trypsin-EDTA (Mediatech, catalog number: 25-051-Cl )
- 0.5 M ethylenediaminetetraacetic acid (EDTA) pH 8.0 (Fisher Scientific, catalog number: BP2482-500 )
- 10% sodium dodecyl sulfate (SDS) (Mediatech, catalog number: 46-040-Cl )
- Proteinase K (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EO0491 )
- Ribonuclease A, DNase free (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EN0531 )
- Potassium acetate (Fisher Scientific, catalog number: P171-500 )
- Isopropanol (Fisher Scientific, catalog number: A416-500 )
- Ethanol 70% (Fisher Scientific, catalog number: BP82011 )
- Sodium hydroxide (NaOH) (Fisher Scientific, catalog: S318-500 )
- SYBR Green PCR Master mix (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4309155 )
- Magnesium chloride (MgCl2) (Fisher Scientific, catalog number: BP214-500 )
- Sucrose (Fisher Scientific, catalog number: S3-500 )
- Tris-HCl buffer pH 7.4 (Lonza, catalog number: 51237 )
- Potassium chloride (KCl) (Fisher Scientific, catalog number: P330-500 )
- NP-40 10% solution (Thermo Fisher Scientific, catalog number: 28324 )
- 1 M MgCl2 (see Recipes)
- 2.5 M sucrose (see Recipes)
- TMK (see Recipes)
- TMK + 1% NP-40 (see Recipes)
- S1 buffer (see Recipes)
- S2 buffer (see Recipes)
- Cell lysis buffer (see Recipes)
Equipment
- Ventilated microcentrifuge (Fisher Scientific, model: accuSpinTM Micro 17R, catalog number: 13-100-676 )
- Gilson PIPETMAN Classic pipets (Gilson, model: P1000, catalog number: F123602 )
- Gilson PIPETMAN Classic pipets (Gilson, model: P20, catalog number: F123600 )
- Gilson PIPETMAN Classic pipets (Gilson, model: P200, catalog number: F123601 )
- Gilson PIPETMAN Classic pipets (Gilson, model: P2, catalog number: F144801 )
- Water bath (Fisher Scientific, model: FS-14 )
- Vortexer (Fisher Scientific, catalog number: 02-216-108 )
- ABI Prism 7900HT sequence detection system (PE-Applied Biosystems) (Thermo Fisher Scientific, Applied BiosystemsTM, model: ABI PRISMTM 7900HT )
Procedure
- NP-40 fractionation
- Wash plate 2 x with 2 ml PBS per well. Plate should have 5-10 x 106 cells at 90-95% confluence.
- Trypsinize cells (500 μl/well) and add 1 ml/well of media to neutralize. Combine the 6 wells of each plate. Can use a single 10 cm dish at the same confluence of 90-95% with 3-6 x107 cells.
- Spin cells at 4 °C, 100 x g for 5 min.
- Resuspend cells with 1 ml PBS.
- Pellet cells at 100 x g for 4 min at 4 °C.
- Resuspend cells in 200 μl TMK.
Note: For qPCR analysis of whole cell extraction, you can take out 50 μl and extract the DNA before proceeding to Procedure B. - Add 150 μl of TMK +1% NP-40.
- Incubate on ice for 5 min with occasional gentle vortexing.
- Pellet at 228 x g for 5 min at 4 °C.
- Remove the supernatant without disturbing the pellet–This is the Cytoplasmic fraction.
- Suspend the stripped nuclear pellet in 500 ml of S1 buffer.
- Layer the S1 suspended pellets over 500 ml of S2 buffer.
- Centrifuge at 1,430 x g for 5 min at 4 °C.
- Aspirate supernatant off.
- Resuspend the nuclei in 300 μl PBS.
Note: At this point, all samples can be stored at 4 °C overnight before proceeding to Procedure B.
- Wash plate 2 x with 2 ml PBS per well. Plate should have 5-10 x 106 cells at 90-95% confluence.
- DNA extractions
- Add 600 μl ice-cold cell lysis buffer to the cell extracts. Expect SDS precipitation with most extractions; it will not affect the end result. Vortex thoroughly for 30 sec.
- Add 3 μl of Proteinase K solution (20 mg/ml). Incubate DNA sample at 55 °C for 3-16 h. The time is dependent on how much SDS precipitation is noticed on the samples and cell line used. A rule of thumb is the more SDS precipitation and larger cells tend to require more incubation time.
- Shear DNA by drawing repeatedly (15 x) through a 21 G needle, avoiding excessive foaming and loss by aerosolization.
- Allow cooling to RT, then add 3 μl of 4 mg/ml DNAse-free Ribonuclease A. Digest for 15-60 min at 37 °C.
Note: At this point, all samples can be stored at 4 °C overnight before proceeding to step B5. - Cool to RT. Add 200 μl 5 M potassium acetate; vortex for 20 sec.
- Pellet precipitated protein/SDS by centrifugation at 4 °C for 3 min at maximum RPM.
- Transfer solution to a fresh tube with 600 μl isopropanol. Mix by inversion. Spin at maximum RPM for 3 min at RT.
- Remove supernatant and wash DNA with 600 μl 70% EtOH. Spin for 3 min at maximum RPM to sediment the pellet. Aspirate the ethanol. Repeat with a 1-min spin and aspirate residual EtOH.
- Air-dry for 15 min.
- Redissolve the pellet in 100 μl 8 mM NaOH or 10 mM Tris. This may be slow–an incubation at 37 °C for 15 min is recommended.
- The DNA may now be stored at -20 °C without incident.
- Add 600 μl ice-cold cell lysis buffer to the cell extracts. Expect SDS precipitation with most extractions; it will not affect the end result. Vortex thoroughly for 30 sec.
- Assessing efficiency of the NP-40 fractionation
To assess the accuracy of the nucleo-cytoplasmic separation, qPCR can be utilized using nuclear, mitochondrial and whole cell extract samples such as example below.- DNA samples were diluted 1 to 10 in dH2O and incubated at 98 °C for 20 min followed by immediate cooling on ice.
- Each PCR reaction contained 7.5 μl SYBR Green PCR Master mix (Fermentas), 2 μl of template, and 1 μM of each primer (Table 1) in a 15 μl reaction volume.
Table 1. Primers for DNA amplification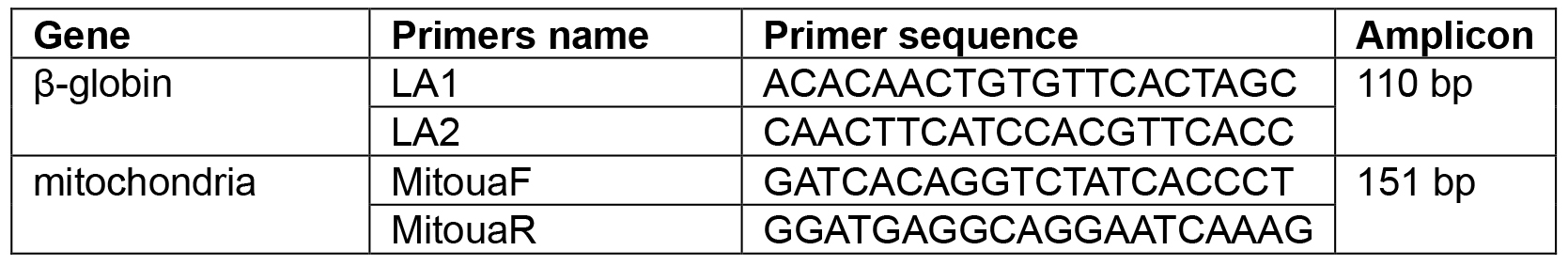
- QPCR was performed in triplicate for each sample using an ABI Prism 7900HT sequence detection system (PE-Applied Biosystems) for amplification and detection. PCR conditions were as follows:

- DNA samples were diluted 1 to 10 in dH2O and incubated at 98 °C for 20 min followed by immediate cooling on ice.
Data analysis
- The accuracy of the NP-40 nucleo-cytoplasmic fraction can be verified by comparing the amount of mitochondrial DNA copies detected in the nuclear fraction and the amount of β-globin DNA copies in the cytoplasmic fraction with the respective copies found in the total cell lysate. The data is analyzed by delta method which requires an average threshold cycle and standard deviation. Hence,
Ct = threshold cycle, a = nuclear or cytoplasmic sample, b = whole cell lysate sample:
- The yield recovered is compared to cytoplasmic contamination in nuclear fractions. Figure 1 adapted from Galvis (2014) is an example of the data analysis graphs that can be composed with the obtained data. The ratio of mitochondrial DNA copies in the nuclear fractions to mitochondrial DNA copies in the whole cell lysate. The example demonstrates a successful fractionation. The nuclear fraction has a 1:1 ratio of nuclear DNA with the whole cell extract; while at the same time there is minimal mitochondrial DNA in comparison to the whole cell extract. The cytoplasmic fraction contains minimal nuclear DNA in comparison to the whole cell extract while it contains an almost 1:1 ratio of mitochondrial DNA with the whole cell extract.
- Note that the recommended inclusion data is that all triplicates must be within one cycle apart from each other and all obtained cycles must be smaller than the negative control that need to be included in the qPCR runs.
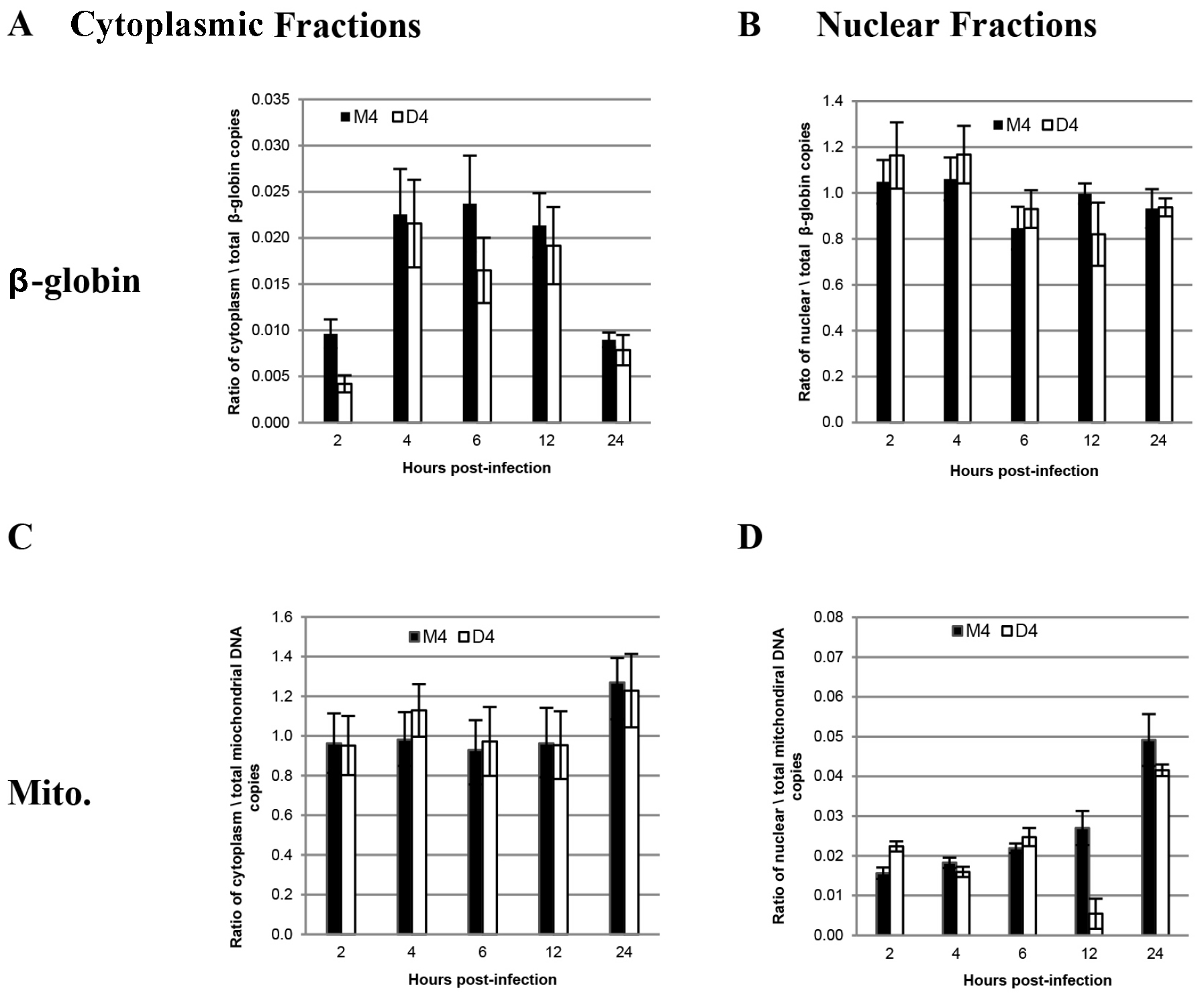 Figure 1. Sample data analysis of fractionation protocol. This is a ratio of mitochondrial and β-globin DNA in nuclear and cytoplasmic fractions vs. total cell lysates. GHOST-R5X4 cells were transfected either with DBR1 shRNA pHyper-D4 (D4) or DBR1 shRNA triple mismatch, pHyper-M4 (M4). Two to twenty-four hours later, the cells were either fractionated into nuclear and cytoplasmic fractions or lysed to prepare whole cell extracts. DNA was isolated for qPCR to evaluate the success of fractionation. A. β-Globin primers were utilized to assay the exclusion of nuclear contamination in the cytoplasmic fraction in contrast to whole cell extracts. The graph illustrates the ratio of β-globin copies in the cytoplasmic fractions to β-globin copies in the whole cell lysates. B. β-Globin primers were utilized to assay concentration of nuclear fractions compared to whole cell extracts. The graph illustrates the ratio of β-globin copies in the nuclear fractions to β-globin copies in the whole cell lysate. C. Mitochondrial primers were used to assay concentration of cytoplasmic fraction compared to whole cell extracts. The graph illustrates the ratio of mitochondrial DNA copies in the nuclear fractions to β-globin copies in the whole cell lysates. D. Mitochondrial primers were utilized to assay the exclusion of mitochondrial contamination in the nuclear extractions.
Figure 1. Sample data analysis of fractionation protocol. This is a ratio of mitochondrial and β-globin DNA in nuclear and cytoplasmic fractions vs. total cell lysates. GHOST-R5X4 cells were transfected either with DBR1 shRNA pHyper-D4 (D4) or DBR1 shRNA triple mismatch, pHyper-M4 (M4). Two to twenty-four hours later, the cells were either fractionated into nuclear and cytoplasmic fractions or lysed to prepare whole cell extracts. DNA was isolated for qPCR to evaluate the success of fractionation. A. β-Globin primers were utilized to assay the exclusion of nuclear contamination in the cytoplasmic fraction in contrast to whole cell extracts. The graph illustrates the ratio of β-globin copies in the cytoplasmic fractions to β-globin copies in the whole cell lysates. B. β-Globin primers were utilized to assay concentration of nuclear fractions compared to whole cell extracts. The graph illustrates the ratio of β-globin copies in the nuclear fractions to β-globin copies in the whole cell lysate. C. Mitochondrial primers were used to assay concentration of cytoplasmic fraction compared to whole cell extracts. The graph illustrates the ratio of mitochondrial DNA copies in the nuclear fractions to β-globin copies in the whole cell lysates. D. Mitochondrial primers were utilized to assay the exclusion of mitochondrial contamination in the nuclear extractions.
Notes
- The data presented in Procedure C were the unpublished controls for experiments denoted in Galvis et al., 2014. Debranching enzyme 1 (DBR1) in our experiments has been implicated in reverse transcription of the HIV-1 genome.
- In Procedure A step 12, it is very important not to disturb the pellet and hence it is better to leave some of the supernatant behind. The purpose of the subsequent steps is to remove the remaining mitochondrial DNA.
- In our experience, the resuspension of the DNA with 8 mM NaOH has given us better overall recovery yield for downstream applications in comparison to 10 mM Tris. However, if the experiments desired only need for qPCR purposes only and not sequencing then 10 mM Tris is sufficient for resuspension of the final pellet.
Recipes
- 1 M MgCl2 (500 ml)
Add 101.65 g MgCl2·6H2O into 500 ml ddH2O
Store at room temperature - 2.5 M sucrose
8.55 g of sucrose
Up to 10 ml with ddH2O
Need to heat to 37 °C in order to have sucrose fully dissolve
Store at room temperature - TMK (10 ml; store in 4 °C)

- TMK + 1% NP-40 (2 ml)
1.8 ml TMK
200 μl 10% NP-40
Store in 4 °C - S1 buffer
0.25 M sucrose
10 mM MgCl2
- S2 buffer
0.35 M sucrose
0.5 mM MgCl2
Store at 4 °C
- Cell lysis buffer
10 mM Tris-HCl, pH 7.4
0.1% SDS
1 mM EDTA
Store at 4 °C
Acknowledgments
This work was supported in part by grant ID07-I-124 from the California HIV-AIDS Research Program to David Camerini. Support of the UCI Center for Virus Research, UCI Cancer Research Institute and the Chao Family Comprehensive Cancer Center are acknowledged. Alvaro Galvis was supported by the UCI Medical Scientist Training Program, grant T32-GM08620. This protocol is adapted and modified from the original in Sambrook and Russell, 2001.
References
- Galvis, A. E. (2014). An RNA lariat intermediate in HIV-1 cDNA synthesis. ProQuest 3615193.
- Galvis, A. E., Fisher, H. E., Nitta, T., Fan, H. and Camerini, D. (2014). Impairment of HIV-1 cDNA synthesis by DBR1 knockdown. J Virol 88(12): 7054-7069.
- Sambrook, J. and Russell, D. W. (2001). Molecular cloning: a laboratory manual. CSHL Press.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Galvis, A. E., Fisher, H. E. and Camerini, D. (2017). NP-40 Fractionation and Nucleic Acid Extraction in Mammalian Cells. Bio-protocol 7(20): e2584. DOI: 10.21769/BioProtoc.2584.
Category
Molecular Biology > DNA > DNA extraction
Cell Biology > Organelle isolation > Nuclei
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









