- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Densities of Bacterial Endosymbionts of Insects by Real-time PCR
Published: Vol 7, Iss 19, Oct 5, 2017 DOI: 10.21769/BioProtoc.2566 Views: 10593
Reviewed by: Filipa VazModesto Redrejo-RodriguezAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol to Mine Unknown Flanking DNA Using PER-PCR for Genome Walking
Zhou Yu [...] Haixing Li
Feb 20, 2025 1498 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views
Abstract
Increased attention has been paid to the endosymbiotic bacteria of insects. Because most insect endosymbionts are uncultivable, quantitative PCR (qPCR) is a practical and convenient method to quantify endosymbiont titers. Here we report a protocol for real-time qPCR based on SYBR Green I fluorescence as well as some tips to prevent possible pitfalls.
Keywords: BacteriaBackground
Insects often harbor bacterial symbionts of various taxa in their bodies. Such bacterial symbionts (endosymbiotic bacteria) attract great attention because of their profound effects on the host insect. Some bacteria provide essential nutrition to their hosts (Baumann et al., 1995), some confer resistance against parasites (Oliver et al., 2003; Hedges et al., 2008), and some even manipulate reproduction or sex determination of their hosts for their own benefit (Werren et al., 2008; Kageyama et al., 2012). Because most insect endosymbionts are uncultivable, quantitative PCR (qPCR) is a practical and convenient method to quantify endosymbiont titers (Simoncini et al., 2001), possibly complemented by other visualization methods, such as fluorescence in situ hybridization (FISH) (Koga et al., 2009) and/or electron microscopy.
Materials and Reagents
Note: Reagents differ depending on the qPCR equipment. Here I describe a protocol for absolute quantification using LightCycler® 480 (Roche). For each reaction, two or more technical replicates are strongly recommended.
- Pipette tips:
10-μl tips (e.g., Fukaekasei and Watson, catalog number: 110-201C )
200-μl tips (Fukaekasei and Watson, catalog number: 1201-705C )
1,000-μl tips (Fukaekasei and Watson, catalog number: 110-706C )
To reduce the risk of contamination, use of the filtered tips, e.g.,
10-μl tips (Fukaekasei and Watson, catalog number: 1251-204CS )
200-μl tips (Fukaekasei and Watson, catalog number: 1252-703CS )
1,000-μl tips (Fukaekasei and Watson, catalog number: 124-1000S )] is preferable - Centrifuge tubes of 1.5 ml (e.g., Fukaekasei and Watson, catalog number: 131-7155C )
- Centrifuge tubes of 50 ml [e.g., Falcon 50-ml conical centrifuge tubes (Corning, Falcon®, catalog number: 352070 )]
- LightCycler® 480 Multiwell Plate 96, white (Roche Molecular Systems, catalog number: 04729692001 , sealing foils included)
- Template DNA to be measured (genomic DNA extracted from insects)
- Template DNA for standard (PCR products or inserted plasmid DNA with known concentration)
- LightCycler® 480, buffer 1 (2x Master mix) (Roche Molecular Systems, catalog number: 04707516001 )
- LightCycler® 480, buffer 2 (H2O) (Roche Molecular Systems, catalog number: 04707516001 )
- Forward and reverse primers
- Reaction mix (see Recipes)
Equipment
- Spectrophotometer (Bio-Rad Laboratories, model: SmartSpecTM 3000 )
- LightCycler® 480 System (Roche Molecular Systems, model: LightCycler® 480 ) connected to a computer
- Centrifuge for 96-well plate (e.g., Kubota, model: PlateSpin II )
- Pipettes [e.g., PIPETMAN® P10 (Gilson, catalog number: F144802 ), P20 (Gilson, catalog number: F123600 ), P100 (Gilson, catalog number: F123615 ), P200 (Gilson, catalog number: F123601 ), or P1000 (Gilson, catalog number: F123602 )]
- Laminar flow cabinet
Software
- LightCycler® 480 Software, version 1.5.1 (Roche Molecular Systems)
- Primer3Plus free online software (http://primer3plus.com/)
Procedure
- Design primer
Use a single-copy gene of a target bacterium for primer design (avoid using ribosomal genes because of the possible presence of operonic copies). For example, the bacterial gene RpoB (Case et al., 2007) is considered suitable. If necessary, design primers for the host (insect) gene to standardize the bacterial titers. Use a single-copy nuclear gene from an autosome (mitochondrial genes are not recommended).
Generally, primers which are 18-23 mer in length are used. The product size should be 80-300 bp (optimum: 80-150 bp). The desired melting temperature (Tm) is 55-65 °C, with a maximum difference of 3 °C in the Tm of the two primers. It is strongly recommended using a software, such as Primer3Plus free online software (http://primer3plus.com/), to design primers.
Some of the primers that have been successfully used to detect insect endosymbionts are given in Table 1.
Table 1. Examples of primers for endosymbiotic bacteria that have been successfully used for qPCR
- Prepare DNA template
Extract DNA from the whole body or a particular tissue/organ of the insect. Usually, 50-100 ng of DNA per reaction is needed. The use of purified DNA, such as that extracted using the phenol chloroform method (Green and Sambrook, 2012) or a commercial kit, such as DNeasy® Blood & Tissue Kit (QIAGEN), is recommended. The concentration of the extracted DNA can be measured using a spectrophotometer. Confirm that A260/A280 is approximately 1.8 and A260/A230 is > 1. If possible, DNA extracted from insects treated with antibiotics should also be used as a control, because this will indicate whether the amplified DNA is derived from the bacterial or the host genome (see Note 1 for details). - Prepare standard DNA
For absolute quantification, DNA samples with known copies of target genes are required. The DNA samples could be PCR products or inserted plasmids. The number of copies can be estimated by mass concentration measured using a spectrophotometer with Avogadro constant NA = 6.022 x 1023 copies/mol, assuming that the molecular weight of 1 bp dsDNA is approximately 660 g/mol. Use a serial dilution of standard DNA, such as 108, 107, 106, 105, 104, and 103 copies. - Prepare the reaction mix (see Recipes), vortex for 5 sec, and dispense 20 μl into each well.
Apply 5 μl of DNA template (sample or standard DNA) or buffer 2 (negative control) (Figure 1).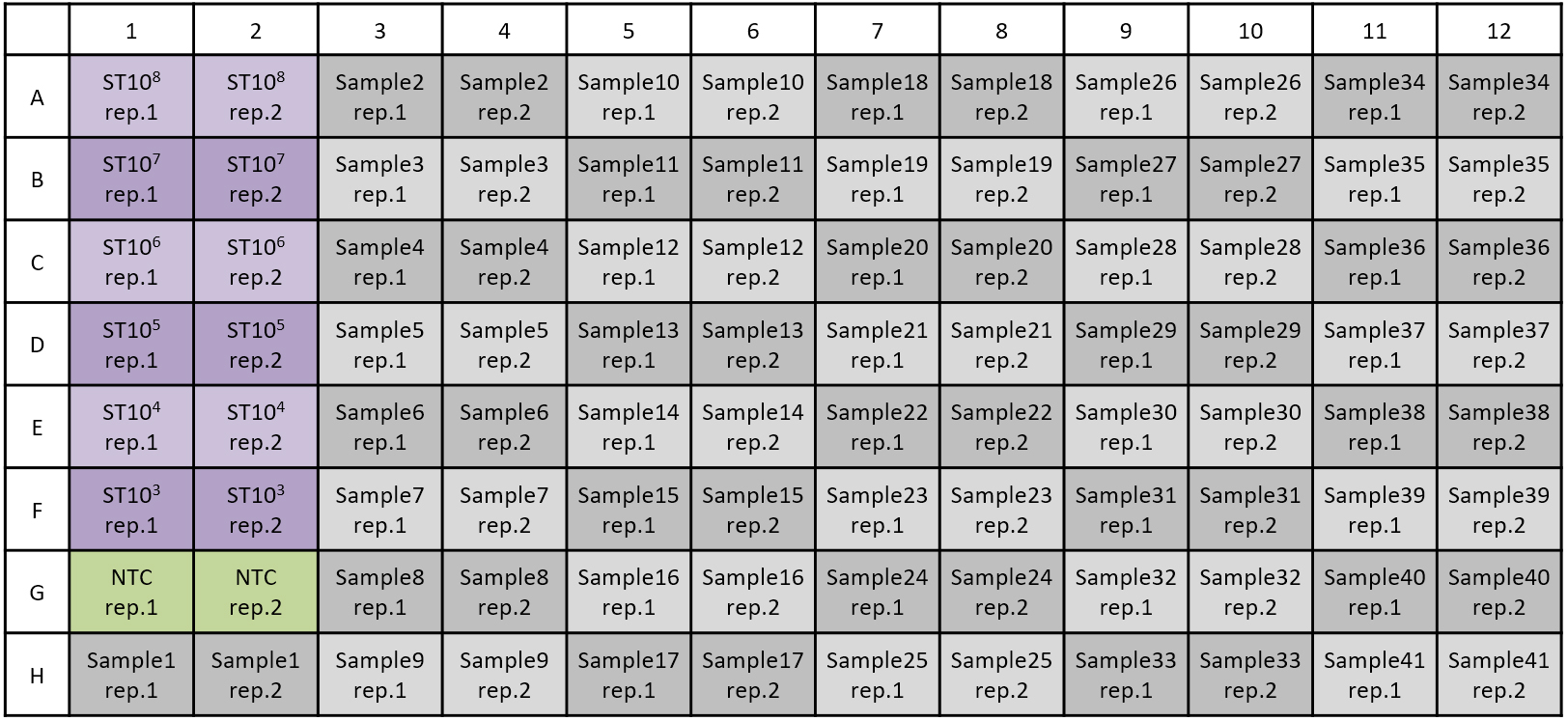
Figure 1. Example of sample sheet. ST: Standard DNA with known concentration, NTC: Non-template control (H2O). When working with two replicates, 41 samples can be measured. - Seal the multiwell plate with sealing foil and centrifuge at 900 x g for 30 sec.
- A typical PCR program may be as follows:
- Initial denaturation for 30 sec at 95 °C.
- Denaturation for 5 sec at 95 °C.
- Annealing for 10-30 sec at X °C (X depends on Tm).
- Extension for 10-20 sec (depends on product length, 1 min kb-1) at 72 °C.
- Return to step (b) for 40 cycles.
- Melting at 60 °C-95 °C.
- Initial denaturation for 30 sec at 95 °C.
- Denaturation for 5 sec at 95 °C.
- Annealing and extension for 10-30 s at X °C (X depends on Tm)
- Return to step (b) for 40 cycles.
- Melting at 60 °C-95 °C.
- Data acquisition should be performed at the extension step in each cycle (necessary for generating an amplification curve) and continuously performed at the final melting (necessary for generating a melting curve; Figure 2).
- When preparing mixtures, special care should be taken to avoid contamination. It is recommended to use particular pipettes for this purpose in a clean environment, such as in a laminar flow cabinet.
- A conserved bacterial gene can be chosen if it is not necessary to classify the bacteria. If the focus is on a specific family, genus, or species, the target gene should be accordingly chosen. Genes that have multiple copies in the genome, such as many outer membrane protein genes, should be avoided. Notably, strain-specific and SNP-containing genes should be avoided because it may compromise the experiment.
- When designing primers, the specificity of the primer to the target gene is important. A BLAST search for possible existence of similar nontarget sequences would be helpful for selecting specific primers. It is also desirable that the products be approximately of the same size.
- Initial denaturation for 30 sec at 95 °C.
Data analysis
After each reaction, check the melting curve to confirm that the correct products have been amplified. Samples with cycle threshold (Ct) value of > 35 should be considered uninfected or infected at an undetectable level.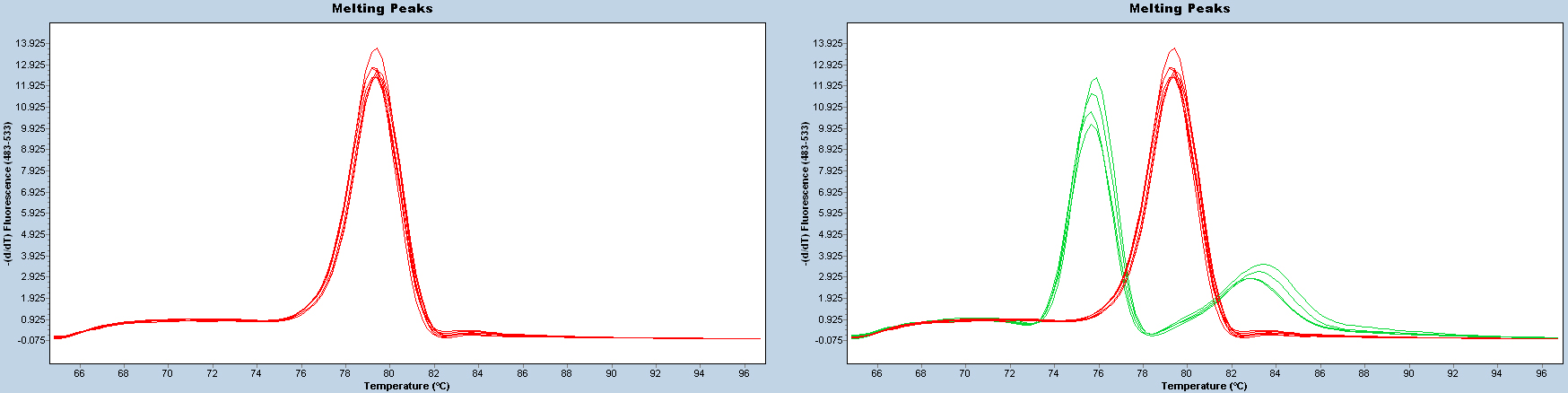
Figure 2. Melting curves. Melting curves should be uniform (red lines on the left panel). Two or more patterns (e.g., red and green lines on the right panel) indicate that PCR products are different.
Notes
- For detection of any bacterial endosymbiont, it is risky to rely only on PCR and qPCR because of possible pseudo-positives (although most can be identified using melting curves) or the amplification of fragments arising due to horizontal gene transfer (HGT). It is known that many insect species harbor bacterial genome fragments of various sizes in their chromosomes (Dunning Hotopp et al., 2007). Therefore, it is desirable to use multiple target genes to reduce the probability of targeting an HGT gene from the insect genome. If possible, visualize the targeted bacterium using FISH before or after examining the bacterial titers by qPCR. It is helpful to also test insects treated with antibiotics (tetracycline hydrochloride is effective for almost all bacterial species) to demonstrate whether this eliminates/decreases the endosymbiont population.
- To standardize the bacterial titers, it is desirable to use the mass concentration of the total genomic DNA as a denominator. However, this may lead to an imprecise estimate of bacterial density because the contribution of bacterial DNA to the total genomic DNA may be significant. For example, Wolbachia pipientis (genome size: ca. 1.2 Mb) whose density is relatively low compared with other obligate endosymbionts, usually yields approximately 10-50 times the number of copies as that of the insect nuclear DNA (Goto et al., 2006). Because the insect genome size is generally 100-300 Mb, bacterial DNA may account for as much as 3.3%-50% of the total genomic DNA, which is too large a proportion to be ignored.
- For standardization of bacterial titers, mitochondrial sequence copies are not recommended, because the density of mitochondria can change according to environment, host physiology, and use of antibiotics. Notably, tetracycline treatment influences mitochondrial metabolism and mtDNA density two generations after treatment in Drosophila simulans (Ballard and Melvin, 2007).
Recipes
- Reaction mix
In each well (25-μl reaction), add the following reagents:
Buffer 1 (2x buffer), 12.5 μl
Buffer 2 (H2O), 2.5 μl
Primer A (10 μM), 2.5 μl
Primer B (10 μM), 2.5 μl
Usually, for one plate (96 wells), a premix can be prepared in a 50-ml tube by adding reagents in the following volumes:
Buffer 1 (2x buffer), 1,250 μl
Buffer 2 (H2O), 250 μl
Primer A (10 μM), 250 μl
Primer B (10 μM), 250 μl
Acknowledgments
This work was supported by KAKENHI (16K08106), Japan.
References
- Ballard, J. W. and Melvin, R. G. (2007). Tetracycline treatment influences mitochondrial metabolism and mtDNA density two generations after treatment in Drosophila. Insect Mol Biol 16(6): 799-802.
- Baumann, P., Baumann, L., Lai, C. Y., Rouhbakhsh, D., Moran, N. A. and Clark, M. A. (1995). Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol 49: 55-94.
- Case, R. J., Boucher, Y., Dahllöf, I., Holmström, C., Doolittle, W. F. and Kjelleberg, S. (2007). Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol 73: 278-288.
- Caspi-Fluger, A., Inbar, M., Mozes-Daube, N., Mouton, L., Hunter, M. S. and Zchori-Fein, E. (2011). Rickettsia ‘in’ and ‘out’: two different localization patterns of a bacterial symbiont in the same insect species. PLoS One 6(6): e21096.
- Dunning Hotopp, J. C., Clark, M. E., Oliveira, D. C., Foster, J. M., Fischer, P., Munoz Torres, M. C., Giebel, J. D., Kumar, N., Ishmael, N., Wang, S., Ingram, J., Nene, R. V., Shepard, J., Tomkins, J., Richards, S., Spiro, D. J., Ghedin, E., Slatko, B. E., Tettelin, H. and Werren, J. H. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317(5845): 1753-1756.
- Goto, S., Anbutsu, H. and Fukatsu, T. (2006). Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl Environ Microbiol 72(7): 4805-4810.
- Green, M. R. and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual, Fourth Edition (3-Volume Set). Cold Spring Harbor Laboratory pp: 1890.
- Hayashi, M., Watanabe, M., Yukuhiro, F., Nomura, M. and Kageyama, D. (2016). A nightmare for males? a maternally transmitted male-killing bacterium and strong female bias in a green lacewing population. PLoS One 11: e0155794.
- Hedges, L. M., Brownlie, J. C., O’Neill, S. L. and Johnson, K. N. (2008). Wolbachia and virus protection in insects. Science 322(5902): 702.
- Kageyama, D., Anbutsu, H., Watada, M., Hosokawa, T., Shimada, M. and Fukatsu, T. (2006). Prevalence of non-male-killing spiroplasma in natural populations of Drosophila hydei. Appl Environ Microbiol 72: 6667-6673.
- Kageyama, D., Narita, S. and Watanabe, M. (2012). Insect sex determination manipulated by their endosymbionts: incidences, mechanisms and implications. Insects 3(1): 161-199.
- Koga, R., Tsuchida, T. and Fukatsu, T. (2009). Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl Entomol Zool 44(2): 281-291.
- Narita, S., Nomura, M. and Kageyama, D. (2007). Naturally occurring single and double infection with Wolbachia strains in the butterfly Eurema hecabe: transmission efficiencies and population density dynamics of each Wolbachia strain. FEMS Microbiol Ecol 61: 235-245.
- Oliver, K. M., Russell, J. A., Moran, N. A. and Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100: 1803-1807.
- Simoncini, L., Casiraghi, M., Bazzocchi, C., Sacchi, L., Bandi, C. and Genchi, C. (2001). Real-time PCR for quantification of the bacterial endosymbionts (Wolbachia) of filarial nematodes. Parassitologia 43(4): 173-178.
- Watanabe, M., Yukuhiro, F., Maeda, T., Miura, K. and Kageyama, D. (2014). Novel strain of Spiroplasma found in flower bugs of the genus Orius (Hemiptera: Anthocoridae): transovarial transmission, coexistence with Wolbachia and varied population density. Microb Ecol 67: 219-228.
- Werren, J. H., Baldo, L. and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6: 741-751.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kageyama, D. (2017). Quantification of Densities of Bacterial Endosymbionts of Insects by Real-time PCR. Bio-protocol 7(19): e2566. DOI: 10.21769/BioProtoc.2566.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > in vivo model > Bacterium
Molecular Biology > DNA > PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











