- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Expression and Purification of the Hsp90-Cdc37-Cdk4 Kinase Complex from Saccharomyces cerevisiae
Published: Vol 7, Iss 19, Oct 5, 2017 DOI: 10.21769/BioProtoc.2563 Views: 9456
Reviewed by: Arsalan DaudiYann Simon GallotAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Protocol to Purify Human Mediator Complex From Freestyle 293-F Cells
Hui-Chi Tang [...] Ti-Chun Chao
Feb 20, 2025 2148 Views

Cell-Sonar, an Easy and Low-cost Method to Track a Target Protein by Expression Changes of Specific Protein Markers
Sabrina Brockmöller [...] Simone Rothmiller
Feb 5, 2025 1657 Views

Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1207 Views
Abstract
Interactions between Hsp90, its co-chaperone Cdc37 and kinases have been biochemically studied for over three decades and have been shown to be functionally important in organisms from yeast to humans. However, formation of a stable complex for structural studies has been elusive. In this protocol we describe expression and purification of Hsp90-Cdc37-Cdk4 kinase protein complex from Saccharomyces cerevisiae utilizing the viral 2A sequences to titrate the three proteins at similar levels.
Keywords: Hsp90Background
Robustly forming complexes between Hsp90 molecular chaperone and its client kinases has proven to be refractory in vitro. Previous work indicated that overexpression of Hsp90’s co-chaperone Cdc37 together with a client kinase in insect Sf9 cells led to a stable complex between Sf9 Hsp90, exogenous Cdc37 and exogenous kinase (Vaughan et al., 2006). However, insect cell culture requires special equipment, is more difficult to genetically manipulate and is significantly slower to both grow and clone into than other well studied expression systems, like bacteria and yeast. Co-expression of the above proteins in E. coli did not yield soluble kinase/stable complex. We reasoned that Saccharomyces cerevisiae would possess the necessary machinery to help fold and facilitate the complex formation and sought to generate the complex between human Hsp90 beta, human Cdc37 and human Cdk4 kinase by co-expressing these proteins in S. cerevisiae. To attain stoichiometric expression of the three proteins, we utilized viral 2A peptides, which allowed transcription of the three proteins on one mRNA with subsequent cleaving at the translation stage. This system has been utilized in human cell lines and in rabbit reticulolysates (Kim et al., 2011; Minskaia and Ryan, 2013), but to our knowledge this is the first utilization of 2A viral expression peptides in S. cerevisiae.
Materials and Reagents
- Generating the co-expression construct
- 100 x 15 mm Petri dishes (Fisher Scientific, catalog number: FB0875713 )
- 15 ml culture tubes (Corning, Falcon®, catalog number: 352051 )
- 1.5 ml microcentrifuge tubes (Fisher Scientific, catalog number: 05-408-129 )
- 0.2 μm filter
- 83nu vector (obtained from Arkin lab)
- GeneArt Strings DNA (Thermo Fisher Scientific)
- NEB Builder Assembly Tool (Online: http://nebuilder.neb.com/)
- Gibson Assembly Cloning Kit (New England Biolabs, catalog number: E5510S )
- DpnI
- PCR Clean up Kit (Promega, catalog number: A9281 )
- Miniprep Kit (Omega Bio-tek, catalog number: D6942-02 )
- Q5 Site-Directed Mutagenesis Kit (New England Biolabs, catalog number: E0554S )
- Bacto-tryptone (BD, BactoTM, catalog number: 211705 )
- Carbenicillin (Gold Bio, catalog number: C-103-100 )
- Yeast extract (BD, BactoTM, catalog number: 212750 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888-10KG )
- Agar (BD, DifcoTM, catalog number: 281210 )
- LB (see Recipes)
- LB agar (see Recipes)
- Expression of the Hsp90/Cdc37/Cdk4
- Disposable spectrophotometer cells (Agilent Technologies, catalog number: 6610018700 )
- JEL1 strain of Saccharomyces cereviase (MAT-alpha, leu2 trp1 ura3-52 prb1-1122 pep4-3 deltahis3::PGAL10-GAL4). Gift from Luke Rice, UT Southwestern
- Frozen-EZ Yeast Transformation II Kit (ZYMO RESEARCH, catalog number: T2001 )
- Galactose (Sigma-Aldrich, catalog number: G0625 )
- Yeast nitrogen base (YNB) (BD, DifcoTM, catalog number: 291940 )
- Glucose (Sigma-Aldrich, catalog number: G8270 )
- CSM-His amino acid mixture (MP Biomedicals, catalog number: 4510-312 )
- Peptone (BD, BactoTM, catalog number: 211820 )
- Yeast extract (BD, BactoTM, catalog number: 212750 )
- Sodium DL-lactate solution (Sigma-Aldrich, catalog number: L1375 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- SD-His (see Recipes)
- YPGL media (see Recipes)
- Disposable spectrophotometer cells (Agilent Technologies, catalog number: 6610018700 )
- Purification of the Hsp90/Cdc37/Cdk4 complex
- 30 ml syringe (BD, catalog number: 309650 ) with 16 G needle (BD, catalog number: 305196 )
- 50 ml conical tubes (Corning, catalog number: 352070 )
- Concentrators, 15 ml, 30 kDa cutoff (EMD Millipore, catalog number: UFC903096 )
- Dialysis tubing, 10 kDa cutoff (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 68100 )
- 12 x 75 mm tubes for fraction collection (Fisher Scientific, catalog number: 14-959-16 )
- FLAG peptide (GenScript, catalog number: RP10586 )
- Protease inhibitors, EDTA free (Roche Diagnostics, catalog number: 11873580001 )
- Ni-NTA Superflow beads (QIAGEN, catalog number: 30450 )
- Anti-FLAG M2 Magnetic Beads (Sigma-Aldrich, catalog number: M8823 )
- TEV protease, prepped in lab, 10 mg/ml, same as (Sigma-Aldrich, catalog number: T4455 )
- Liquid nitrogen
- Bolt 4-12% Bis-Tris Plus Gels (Thermo Fisher Scientific, InvitrogenTM, catalog number: NW04120BOX )
- NuPAGE LDS sample buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0007 )
- NuPAGE MOPS SDS running buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0001 )
- Trizma base (Sigma-Aldrich, catalog number: T1503 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888-10KG )
- Imidazole (Sigma-Aldrich, catalog number: I2399 )
- Magnesium chloride (MgCl2), 1 M stock (Sigma-Aldrich, catalog number: 63069 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Sodium molybdate (NaMoO4) (Sigma-Aldrich, catalog number: 243655 )
- Dithiothreitol (DTT) (Gold Bio, catalog number: DTT100 )
- Lysis buffer (see Recipes)
- Dialysis buffer (see Recipes)
- Gel filtration buffer (see Recipes)
- 30 ml syringe (BD, catalog number: 309650 ) with 16 G needle (BD, catalog number: 305196 )
Equipment
- Generating the co-expression construct
- PCR machine (like Biorad DNA Engine BG96TC)
- 37 °C culture shaker (like New Brunswick Innova 43 series)
- 4 °C and -20 °C fridge (any brand)
- 42 °C heat block (like Thomas Scientific 3661044)
- PCR machine (like Biorad DNA Engine BG96TC)
- Expression of Hsp90/Cdc37/Cdk4
- 2.5 L flasks (Ultra Yield) (Thomson Instrument, catalog number: 931136-B )
- Autoclave
- Centrifuge capable of 3,000 x g utilizing 1 L bottles (like Beckman Coulter, model: Avanti® J20 Series )
- 30 °C culture shaker capable of shaking at 200 rpm 250 ml and 1 L flasks (like Eppendorf, New BrunswickTM, model: Innova® 4200 series)
- UV-Visible spectrophotometer (Agilent Technologies, model: Agilent 8453 )
- 2.5 L flasks (Ultra Yield) (Thomson Instrument, catalog number: 931136-B )
- Purification of the Hsp90/Cdc37/Cdk4 complex
- EmulsiFlex-C3 (Avestin, model: EmulsiFlex-C3 )
- Centrifuge (like Beckman Coulter, model: Avanti® J20 Series )
- High-speed centrifuge tubes (capable of 30,000 x g like Beckman Coulter, catalog number: 357002 )
- Rocker (like Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 88880019 )
- MonoQ 10/100GL column (GE Healthcare, catalog number: 17-5167-01 )
- HiLoad 16/600 Superdex 200pg column (GE Healthcare, catalog number: 28989335 )
- Glass gravity columns (Bio-Rad Laboratories, catalog number: 7372512 )
- ÄKTApurifier system (GE Healthcare, model: ÄKTApurifier system)
- -80 °C fridge (like Eppendorf, New BrunswickTM, model: U570 ULT )
- Assortment of beaker sizes
- Magnetic stand (like EMD Millipore, catalog number: LSKMAGS15 )
- Mini Gel tank (Thermo Fisher Scientific, catalog number: A25977 )
- PowerEase 500 Power Supply (Thermo Fisher Scientific, model: PowerEase® 500 )
- Liquid nitrogen flask (like Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 2129 )
- Spatulas (Cole-Parmer, catalog number: UX-06369-16 )
- EmulsiFlex-C3 (Avestin, model: EmulsiFlex-C3 )
Procedure
- Generating the co-expression construct
- To co-express human Hsp90β, human Cdc37 and human Cdk4 we utilized viral 2A peptides. This way we were able to construct a single plasmid, which had all three proteins in it. (see Figure 1) The exact 2A sequence we used (referred to as P2A) was sourced from Porcine Teschovirus-1 and was GSGATNFSLLKQAGDVEENPGP. The resulting construct was of this arrangement: hCdc37-TEVsite-P2A-hHsp90β-TEVsite-FLAG-P2A-hCdk4-TEVsite-HisTag. The nucleotide sequence which was inserted between hCdc37 and hHsp90 was: CAGAACCTGTACTTTCAGGGCGGATCCGGTGCCACCAACTTTAGCTTGTTGAAGCAGGCTGGAGACGTGGAGGAAAATCCTGGACCC, between hHsp90 and hCdk4 was: CAAAACTTATACTTCCAGGGTGACTATAAGGACGATGACGACAAAGGCTCCGGTGCCACCAACTTCTCATTATTGAAACAGGCCGGTGATGTAGAGGAAAATCCAGGACCT, and after hCdk4 was: CAGAATCTGTATTTTCAGGGACATCATCACCATCACCATTAATGA. The complete sequence above was codon optimized (the regions between proteins were manually checked to be dissimilar to ease cloning). See Appendix for complete insert sequence.
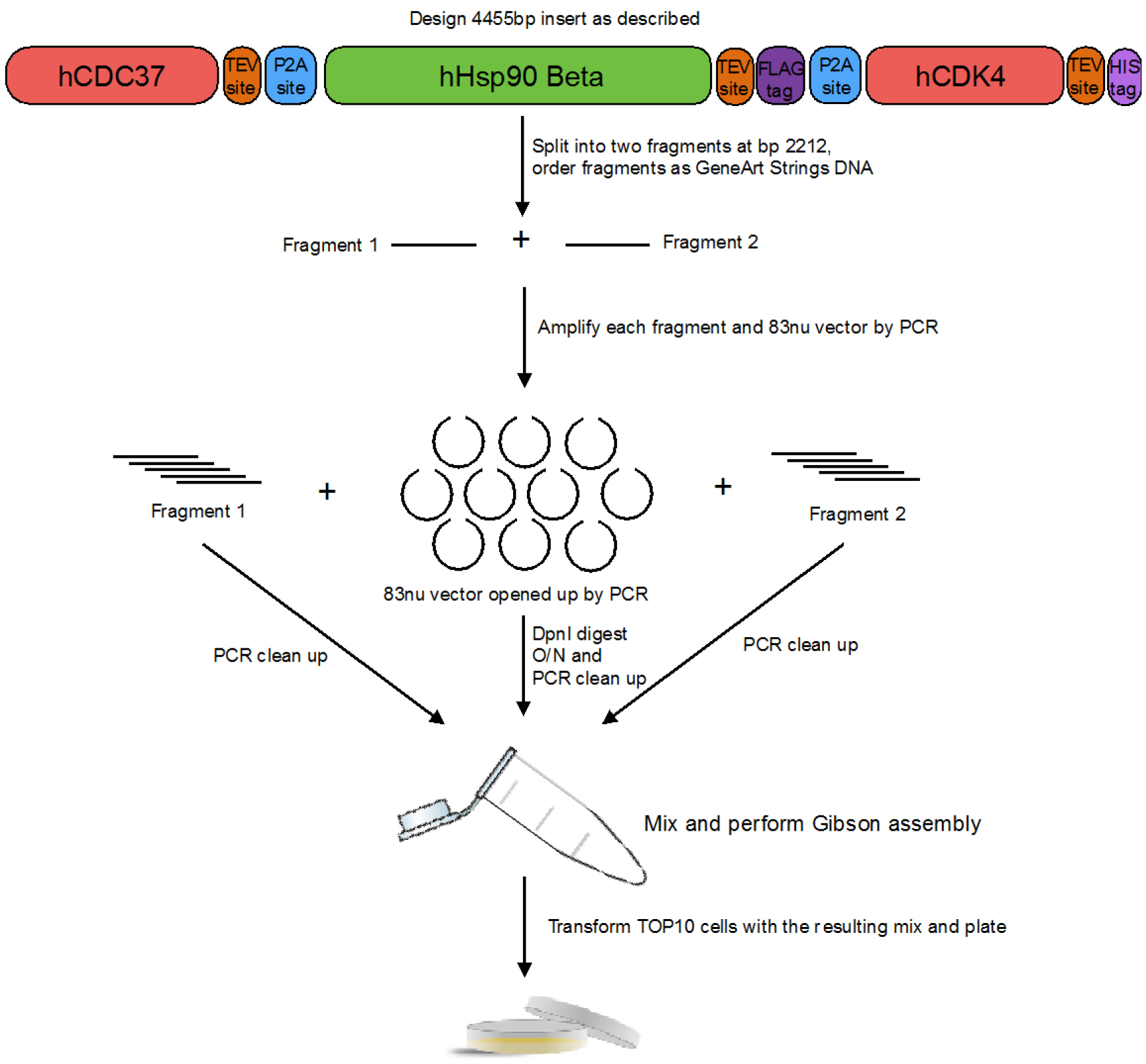
Figure 1. Design and cloning of the construct with Hsp90, Cdc37 and Cdk4 in it - The sequence was then split into two pieces (at nucleotide number 2,212) and both parts were ordered as GeneArt Strings DNA from Thermo Fisher Scientific. Using NEBuilder, primers were designed to amplify the 83nu vector and each of the GeneArt Strings DNA as to clone them using Gibson Assembly Cloning Kit.
- 83nu vector and each of the GeneArt Strings DNA are amplified using a PCR machine, then digested with DpnI at 37 °C overnight (O/N) (~16 h), PCR cleaned up, assembled using Gibson Assembly Cloning Kit and transformed into E. coli TOP10 cells supplied with the kit, which are plated and grown O/N at 37 °C on LB-carbenicillin plates.
- Next day, pick 10 colonies and start a 5 ml culture of LB together with 100 mg/L of carbenicillin with each colony.
- After 24 h growth at 37 °C with shaking, the plasmid DNA is extracted utilizing miniprep kit and sent for sequencing (tiling every 500 bp through the whole gene). In our case, the best clone had one mutation due to synthesis errors, which was corrected utilizing Q5 Site Directed Mutagenesis Kit and the protocol therein.
- The final construct is sequenced and stored in distilled nuclease free water at -20 °C for future use. 4 °C fridge is used for short-term storage of DNA (under a week).
- To co-express human Hsp90β, human Cdc37 and human Cdk4 we utilized viral 2A peptides. This way we were able to construct a single plasmid, which had all three proteins in it. (see Figure 1) The exact 2A sequence we used (referred to as P2A) was sourced from Porcine Teschovirus-1 and was GSGATNFSLLKQAGDVEENPGP. The resulting construct was of this arrangement: hCdc37-TEVsite-P2A-hHsp90β-TEVsite-FLAG-P2A-hCdk4-TEVsite-HisTag. The nucleotide sequence which was inserted between hCdc37 and hHsp90 was: CAGAACCTGTACTTTCAGGGCGGATCCGGTGCCACCAACTTTAGCTTGTTGAAGCAGGCTGGAGACGTGGAGGAAAATCCTGGACCC, between hHsp90 and hCdk4 was: CAAAACTTATACTTCCAGGGTGACTATAAGGACGATGACGACAAAGGCTCCGGTGCCACCAACTTCTCATTATTGAAACAGGCCGGTGATGTAGAGGAAAATCCAGGACCT, and after hCdk4 was: CAGAATCTGTATTTTCAGGGACATCATCACCATCACCATTAATGA. The complete sequence above was codon optimized (the regions between proteins were manually checked to be dissimilar to ease cloning). See Appendix for complete insert sequence.
- Expression of the Hsp90/Cdc37/Cdk4
- The plasmid is transformed into JEL1 (MAT-alpha, leu2 trp1 ura3-52 nprb1-1122 pep4-3 deltahis3::PGAL10-GAL4) yeast strain using Frozen-EZ Yeast Transformation II Kit protocol and plated on SD-His plates.
- After 3 days, pick a colony and inculcate into 100 ml SD-His media in a 250 ml flask and grow at 30 °C O/N shaking at 200 rpm.
- Next day, inoculate 6 x 1 L flasks of YPGL media (autoclaved) each with 10 ml of the O/N culture.
- After about 24 h at 30 °C and 200 rpm (optical density [OD] of 1, measured on the spectrophotometer), add powder galactose to a final concentration of 2% w/v to induce protein expression.
- The culture is pelleted after 6 h of growth at 3,000 x g for 20 min.
- Pellets are scooped up and frozen at -80 °C.
- The plasmid is transformed into JEL1 (MAT-alpha, leu2 trp1 ura3-52 nprb1-1122 pep4-3 deltahis3::PGAL10-GAL4) yeast strain using Frozen-EZ Yeast Transformation II Kit protocol and plated on SD-His plates.
- Purification of the Hsp90/Cdc37/Cdk4 complex
- Resuspend the cell pellets in lysis buffer with protease inhibitors first with spatula, then pass the sample through a 16 G syringe needle (about 160 ml total for 6 L yeast culture).
- Then pass the lysate through EmulsiFlex-C3 machine at 25,000 psi, 5 complete passes for lysis following standard manufacturer’s procedures.
- Clear the lysate by centrifuging at 30,000 x g for 30 min.
- Equilibrate 5 ml bed volume of Ni-NTA beads with 20 bed volumes (100 ml) of lysis buffer at room temperature utilizing a glass gravity column. The supernatant from the centrifuge spin is added to the beads, and then rocked for 1 h at 4 °C in the capped glass gravity column.
- After 1 h incubation, wash the beads with 20 bed volumes of lysis buffer and then elute into 10 bed volumes of lysis buffer + 500 mM imidazole, utilizing a glass gravity column for both steps.
- Then incubate the resulting eluate with pre-equilibrated (10 bed volumes of lysis buffer utilizing the magnetic stand) Anti-FLAG M2 Magnetic Beads (1 ml bed volume) in a 50 ml plastic conical tube for 1 h at 4 °C, rocking.
- Wash the beads with 10 bed volumes of lysis buffer utilizing the magnetic stand and a 15 ml tube, and elute the sample with 3 bed volumes of lysis buffer with 75 μg/ml of FLAG peptide, twice, 25 min each time
- Add TEV protease to the eluent (2.5 mg) and dialyze sample against 4 L of dialysis buffer O/N utilizing dialysis tubing.
- Dilute the sample 1:1 with the dialysis buffer without NaCl and load onto pre-equilibrated MonoQ 10/100GL column on an ÄKTApurifier system.
- After washing out the unbound sample (about 3 CV [column volumes]), a gradient is run up to 1 M NaCl (over 20 CV) with fractionation to elute the bound complex (comes off at about 25% conductivity).
- The fractions are pooled, concentrated and then loaded on HiLoad 16/600 Superdex 200pg column pre-equilibrated in gel filtration buffer.
- The peak fractions (at about 0.5 CV) are pooled, concentrated, flash frozen in liquid nitrogen and stored at -80 °C. At various points (see Figure 2) during the purification aliquots are taken to be run on SDS-PAGE gel (Figure 2).
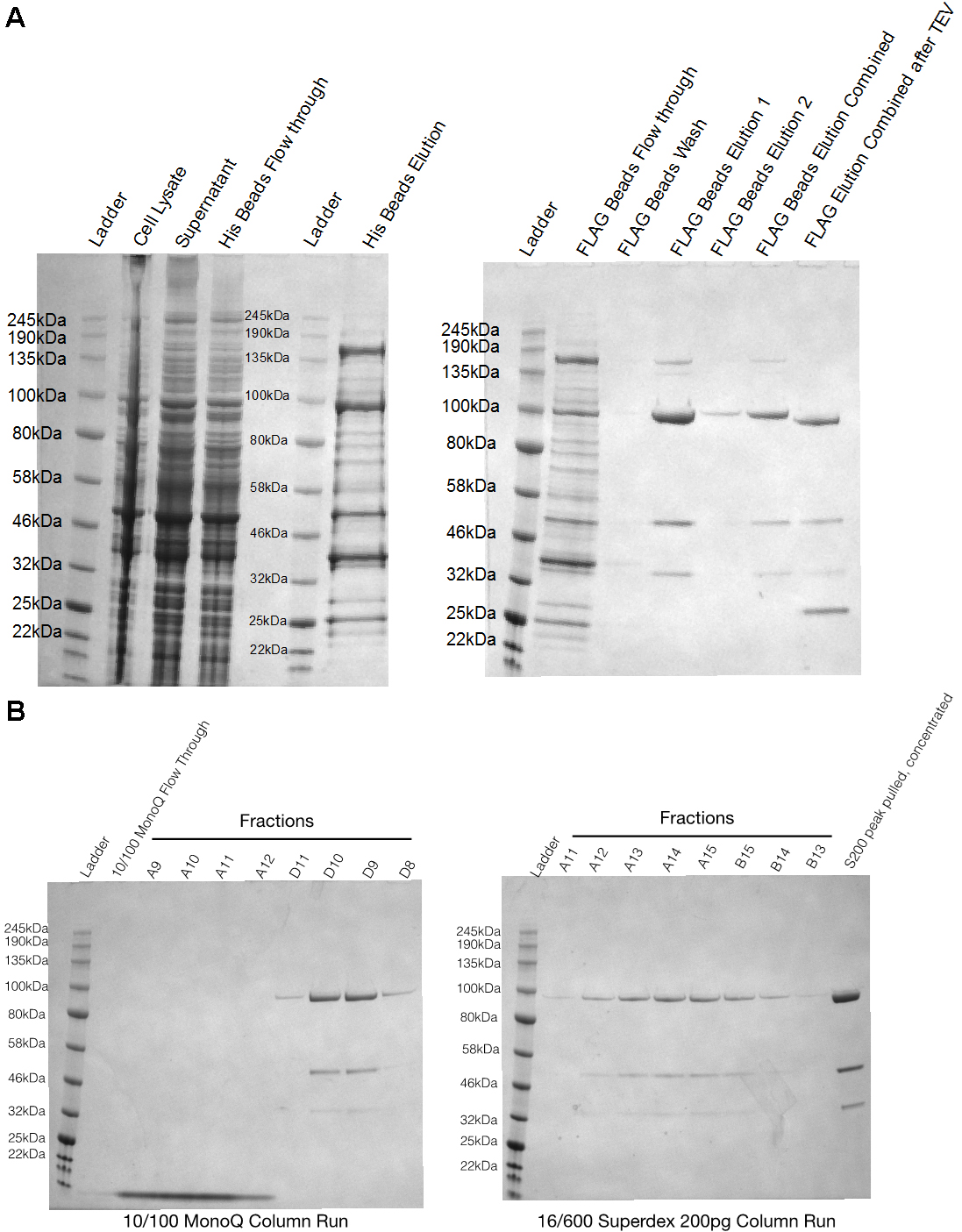
Figure 2. SDS-PAGE gels of Hsp90-Cdc37-Cdk4 purification from yeast. A. SDS-PAGE gels show aliquots at different steps during the purification. Cell lysate–right after step C2 of purification, Supernatant–supernatant at step C3, His beads flow-through–flow-through at step C5, His beads elution–elution at step C5, Flag beads flow through–flow through of flag beads at step C6, Flag beads elution–elution at step 7. B. Two gels are fractions off MonoQ 10/100GL column and size exclusion column runs.
- Resuspend the cell pellets in lysis buffer with protease inhibitors first with spatula, then pass the sample through a 16 G syringe needle (about 160 ml total for 6 L yeast culture).
Data analysis
Sample prepared in this way was subsequently prepared for cryo electron microscopy (cryoEM) followed by data collection and reconstruction. The protocols utilized for these steps are described in great detail in Verba et al., 2016.
Notes
- Scaling up the purification Genscript Anti-DYKDDDDK G1 affinity resin (L00432) was also used successfully with glass gravity column instead of Anti-FLAG M2 Magentic Beads. Use the same amount of beads/proportion of wash/elution buffers.
- This protocol has also been successfully used for expression and purification of Hsp90/Cdc37/Her2 complex.
- Expression of kinase alone, without Hsp90/Cdc37 yields soluble, active kinase.
Recipes
- LB
10 g Bacto-tryptone
5 g yeast extract
10 g NaCl
in 1 L of MilliQ water
Autoclave
If adding antibiotic, like carbenicillin, add 50 mg/ml stock solution (0.2 μm sterile filtered) to the LB cooled to below 50 °C, to a final concentration of 50 mg/L - LB agar
As above with 15 g agar
If adding antibiotic, like carbenicillin, add 50 mg/ml stock solution (0.2 μm sterile filtered) to the LB cooled to below 50 °C, to a final concentration of 50 mg/L - SD-His
6.7 g of yeast nitrogen base
5 g glucose
0.77 g CSM-His amino acid mix
in 1 L of MilliQ water (or 100 ml for 10x)
Sterile filter
For plates proceed as above, but with 15 g of agar per liter - YPGL media
2% peptone
1% yeast extract
3% glycerol
2% lactate
in 1 L of deionized water
Autoclave - Lysis buffer
20 mM Tris pH 7.5
150 mM NaCl
20 mM imidazole
10 mM MgCl2
10 mM KCl
20 mM NaMoO4 - Dialysis buffer
20 mM Tris pH 7.5
100 mM NaCl
10 mM MgCl2
10 mM KCl
20 mM NaMoO4 - Gel filtration buffer
20 mM Tris pH 7.5
150 mM NaCl
10 mM KCl
20 mM NaMoO4
1 mM DTT
Acknowledgments
We thank Akihiko Arakawa (Yokoyama Lab) for sharing their insect cell purification protocol, Yao Fan (Arkin Lab) for the yeast expression vector, and D.A.A. lab members for helpful discussions, specifically Miguel Betegon for the suggestion of using viral 2A peptides and Michelle Moritz for yeast expression strain. Support for this work was provided by PSI-Biology grant U01 GM098254 (to D.A.A.), AACR-BCRF Grant 218084 for Translational Breast Cancer Research (to D.A.A.), The Cabala Family gift (to D.A.A.), HHMI International Student Research Fellowship (to K.V.) and the Howard Hughes Medical Institute (to D.A.A.).
References
- Kim, J. H., Lee, S. R., Li, L. H., Park, H. J., Park, J. H., Lee, K. Y., Kim, M. K., Shin, B. A. and Choi, S. Y. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6(4): e18556.
- Minskaia, E. and Ryan, M. D. (2013). Protein coexpression using FMDV 2A: effect of "linker" residues. Biomed Res Int 2013: 291730.
- Vaughan, C. K., Gohlke, U., Sobott, F., Good, V. M., Ali, M. M., Prodromou, C., Robinson, C. V., Saibil, H. R. and Pearl, L. H. (2006). Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell 23(5): 697-707.
- Verba, K. A., Wang, R. Y., Arakawa, A., Liu, Y., Shirouzu, M., Yokoyama, S. and Agard, D. A. (2016). Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352(6293): 1542-1547.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Verba, K. A. and Agard, D. A. (2017). Protein Expression and Purification of the Hsp90-Cdc37-Cdk4 Kinase Complex from Saccharomyces cerevisiae. Bio-protocol 7(19): e2563. DOI: 10.21769/BioProtoc.2563.
Category
Biochemistry > Protein > Expression
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









