- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Construction and Screening of a Transposon Insertion Library of Yersinia enterocolitica (YeO3-R1)
Published: Vol 2, Iss 15, Aug 5, 2012 DOI: 10.21769/BioProtoc.246 Views: 15092

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Method of Inducible Directed Evolution to Evolve Complex Phenotypes
Ibrahim S. Al’Abri [...] Nathan Crook
Oct 20, 2022 3062 Views

Comprehensive Mapping of EZ-Tn5 Transposon Insertion Sites in Pseudomonas argentinensis SA190 Using RATE-PCR
Büsra Elkatmis [...] Maged M. Saad
Jul 20, 2025 1753 Views

Editing the Serratia proteamaculans Genome Using the Allelic Exchange Method
Ksenia Chukhontseva [...] Ilya Demidyuk
Sep 20, 2025 1390 Views
Abstract
The Mu-transposon system is one of the best characterized transposition systems. Under minimal in vitro set-up, Mu transposition requires only a simple reaction buffer, MuA transposase protein, mini-Mu transposon DNA (donor) and target DNA. The reaction proceeds via initial assembly of the transposition complex that directs transposon integration into target DNA with high efficiency and relatively low target site selectivity. These characteristics make the Mu in vitro transposition technology ideal for the generation of comprehensive mutant DNA libraries usable in a variety of molecular biology applications. This technology has successfully been used for DNA sequencing, functional analyses of plasmid DNA and virus genomes, protein engineering for structure/function and protein-protein interaction studies and generation of gene targeting constructions. When electroporated, the in vitro–assembled Mu transposition complexes can also be used for efficient gene delivery in bacteria, yeasts and mammalian cells. Using this protocol we have identified several mutants where Cat-Mu insertion has interrupted genes involved in lipopolysaccharide (LPS) biosynthesis (Pinta et al., 2012).
Keywords: Yersinia enterocoliticaMaterials and Reagents
- Yersinia enterocolitica strain YeO3-R1 (al-Hendy et al., 1992)
- Cat-Mu transposon (Haapa et al., 1999) or equivalent like Entranceposon Cam-R3 (Thermo Fisher Scientific, Finnzymes, catalog number: F-778 )
- MuA transposase 1,100 ng/μl and MuA storage buffer (Thermo Fisher Scientific, Finnzymes, catalog number: F-750C )
- 99.5% glycerol BDH (catalog number: 24388.320 )
- 10% Triton X-100 (w/v) (F. Hoffmann-La Roche, catalog number: 11332481001 )
- Ultrapure 0.5 M EDTA (pH 8.0) (Life Technologies, Invitrogen™, catalog number: 15575 )
- DTT (Sigma-Aldrich, catalog number: D0632 )
- Centricon YM-100 (100-kDa cutt off) (EMD Millipore, catalog number: UFC210024PL )
- Polyethylene glycol (PEG) 6000 (Merck KGaA, catalog number: 8.07491 )
- NuSieve 3:1 Agarose (25 g) (Lonza, catalog number: 50091 )
- Albumin from bovine serum, BSA (Sigma-Aldrich, catalog number: A7906 )
- Heparin (Sigma-Aldrich, catalog number: H3393 )
- Ficoll PM 400 (Sigma-Aldrich, catalog number: F4375 )
- Bacto-agar, Bacto-tryptone, and Bacto-yeast extract (BD DifcoTM)
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- Tryptic Soya Broth (Oxoid Limited, catalog number: CM0129 )
- Enterocoliticin produced by Y. enterocolitica serotype O: 7, 8 strain 29930 (Strauch et al., 2001)
- MuA storage buffer
- Sodium acetate (NaAc)
- DNA ladder (New England Biolabs)
- Phosphate-buffered saline (PBS) (pH 7.4)
- TGD buffer (see Recipes)
- 5x complex buffer (see Recipes)
- 1 M DTT (see Recipes)
- 20% PEG6000 (w/v) – 2.5 M NaCl (5 ml) (see Recipes)
- SOB medium (1 L) without magnesium (see Recipes)
- 2 M Mg++ stock (see Recipes)
- 2 M glucose (100 ml) (see Recipes)
- SOC medium (100 ml) (see Recipes)
- Luria broth (LB) medium (1 L) (see Recipes)
- LB agar (LA) plates (1 L) (see Recipes)
- Chloramphenicol (Clm) solution (10 ml) (see Recipes)
- Tryptic soya broth (see Recipes)
- Phosphate-buffered saline (PBS) (pH 7.4) (1 L) (see Recipes)
- TAE buffer (see Recipes)
Equipment
- Centrifuges (Sorvall, Heraeus Holding)
- Water bath (Grant)
- Incubator (Termaks)
- Shaker (New Brunswick Scientific)
- Biofotometer (Eppendorf)
- Agarose gel electrophoresis (Bio-Rad Laboratories)
- Gel documentation equipment (Bio-Rad Laboratories)
- Electroporator (Bio-Rad Laboratories, Genepulser II)
- 90 mm Petri dishes (Thermo Fisher Scientific, Sterilin®, catalog number: 101RT )
- 0.1-cm electroporation cuvettes (Bio-Rad Laboratories)
Procedure
- Transposition complex assembly (Lamberg et al., 2002)
- Prepare eight standard assembly reactions using Cat-Mu (1250-bp) transposon as a donor DNA. Cat-Mu (or equivalent) is commercially available. Alternatively, custom-made transposons can be constructed in the context of their carrier plasmids, isolated by BglII digestion and purified by anion exchange chromatography as described (Haapa et al., 1999).
- Mix ddH2O, glycerol, and 5x complex buffer at RT, and transfer the tube on ice. Take appropriate volumes of the reagents according to the chart below.
- Add your donor DNA.
- Dilute the MuA transposase in final concentration of 400 ng/μl with cold MuA storage buffer. Dilute only the amount needed. Keep MuA on ice all the time. Pipet MuA up and down before taking an aliquot or adding into the reaction as it is a sticky protein.
- Add MuA and transfer the tubes directly into 30 °C water bath. Mix thoroughly!
Component Amount (μl) Final ddH2O 16 Final volume 80 μl 99.5% glycerol 40 50% 5x complex buffer 16 1x Cat-Mu (1.1 pmol/907.5 ng/μl) 4 4.4 pmol MuA transposase (400 ng/μl) 4 1,600 ng - Incubate at 30 °C for 4 h.
- Concentrating your complexes
- Following assembly, reactions are pooled, and reaction products are concentrated (~10-fold) and desalted using Centricon YM-100 (100-kDa cut off, Millipore) centrifugal cartridges as described (Pajunen et al., 2005). Pooled assembly reactions are diluted up to 2 ml with TGD buffer, applied on the Centricon, centrifuged 30 min 1,000 x g, washed once with 1 ml of TGD buffer (15 min 1,000 x g), and finally recovered with a reverse spin at 300 x g for 2 min. Typical volumes after Centricon concentrators range from 40 to 70 μl.
- Alternatively, the concentration step can be done using polyethylene glycol (PEG6000) precipitation as described (Paatero et al., 2008).
- Pool your 8 x 80 μl assembly reactions = 640 μl.
- (Optional: take 5 μl sample for transposition complex gel)
- Add 380 μl filter-sterilized 20% PEG6000 – 2.5 M NaCl. Prepare fresh solution each time you concentrate your complexes.
- Incubate (precipitate) on ice 60 min.
- (Optional: take 5 μl sample for gel as above)
- Centrifuge for 60 min 13,000 rpm at 4 °C in a micro centrifuge.
- Discard supernatant carefully without disturbing the pellet.
- Resuspend pellet in 50 μl of TGD buffer. The calculated concentration of the Cat-Mu transposition complex preparation is 580.8 ng/μl.
- Dissolve the pellet on ice for 60 min or overnight. Tap the tube to help the pellet dissolve.
- Preparations are frozen in liquid nitrogen and stored at -80 °C. Transposition complexes can be stored at -80 °C for several months even up to 1 year without loosing activity.
- The assembly and concentration of the transposition complexes can be monitored by agarose/heparin/BSA gels as below (see Figure 1).
- The transpositional activity of the preparation can be measured by an in vitro transposition assay using plasmid DNA as target and introduction of the reaction products into competent E. coli cells (Haapa et al., 1999). Recovery of chloramphenicol-resistant colonies is an indication of functional transposition complexes.
- Transposition complex gel
- Prepare an agarose/heparin/BSA gel (2% NuSieve 3: 1 in 1x TAE, 87 μg/ml heparin, 87 μg/ml BSA).
- Measure 1 g of NuSieve 3: 1 agarose in a 250-ml flask.
- Add 49 ml of ddH2O, and weigh the flask again (should be approx. 49 g).
- Melt agarose in microwave oven (or equivalent).
- Weigh the flask again, and add H2O so that final weight is again 49 g.
- Add 1 ml 50x TAE buffer.
- When the solution is about 55 °C, add 43.5 μl of heparin 100 mg/ml and 109 μl of BSA 40 mg/ml.
- Prepare gel samples. For non-concentrated complexes (5 μl samples), add 0.5 μl of heparin 65 mg/ml, and 1 μl of 25% Ficoll PM 400 (w/v). For concentrated complexes, take 0.5–1 μl samples; add ddH2O up to 5 μl volume, and then heparin/Ficoll as above.
- Run the gel in 1x TAE at 80 V (5 V/cm) for 2 h. Preferably circulate the buffer with a pump.
- Stain the gel with ethidium bromide 0.5 μg/ml in 1x TAE for 60 min. Staining can be prolonged up to overnight if necessary.
- (Optional: Destain the gel in 1x TAE or ddH2O for 20 min.)
- Take a picture (Figure 1).
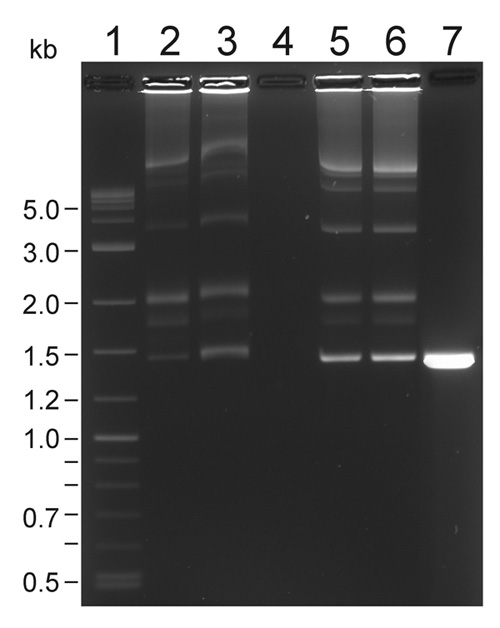
Figure 1. A representative transposition complex gel of Em-Mu (1447-bp) complexes (Pajunen et al., 2005). Lane 1, 300 ng of 2-log DNA ladder. Lane 2, 5 μl sample from pooled assembly reaction (250 ng of donor). Lane 3, 10 μl sample from PEG-precipitation before centrifugation (326 ng). Lane 4, 10 μl sample from supernatant. Lane 5, 0.61 μl sample from concentrated complexes dissolved in TGD buffer (410 ng of donor DNA. Lane 6, 1 μl sample from concentrated complexes (665 ng of donor DNA). Lane 7, unassembled control, 410 ng of donor DNA alone. The ca. 3.5 kb band corrensponds to the complex formation between the ends of a single transposon (C1), see Lamberg et al. (2002) for Cat-Mu complex gel.
- Preparation of electrocompetent cells (Lamberg et al., 2002)
- Inoculate a fresh YeO3-R1 colony into 25 ml SOB medium (without magnesium) in 100 ml flask. Grow the cells overnight with vigorous aeration at 37 °C.
- Dilute 0.5 ml of the overnight culture into 500 ml of SOB in a 3 L flask. Grow for 2-3 h with vigorous aeration at 37 °C until the OD600=0.8.
- Incubate on ice 30 min.
- Divide into three 250 ml bottles. Harvest the cells by centrifugation at 5,000 rpm (2,600 x g) in a Sorvall GSA (or equivalent) rotor for 10 min at 4 °C.
- Wash the cell pellet by resuspending each in 5 ml of sterile ice-cold wash buffer (10% glycerol, 90% distilled water, v/v). Keep the bottles on ice at all times.
- Combine into two bottles.
- Add 200 ml ice-cold wash buffer per bottle, and mix by inverting the bottles several times.
- Centrifuge the cell suspension at 5,000 rpm (2,600 x g) for 15 min at 4 °C and carefully pour off the supernatant as soon as the rotor stops. Cells washed in the Wash buffer do not pellet well. If the supernatant is turbid, increase the centrifugation time.
- Wash the cell pellet a second time by resuspending in 5 ml of sterile ice-cold wash buffer. Add 200 ml wash buffer, and mix by inverting the bottle several times.
- Centrifuge the cell suspension at 5,000 rpm for 15 min.
- Pour off the supernatant and resuspend the cells in the Wash buffer that remains in the centrifuge bottle. Combine into single bottle. Final volume is approximately 2 ml (~1x1011 cells/ml).
- Incubate on ice at least 30 min, preferably 60 min.
- Cells can be used immediately or can be frozen in aliquots. For some strains like YeO3-R1 fresh cells yield better efficiencies.
- For freezing divide 60 μl (or 120 μl) in sterile cold micro centrifuge tubes. Flash-freeze in liquid nitrogen and store at -80 °C.
- Preparation transposon insertion library (Pinta et al., 2012)
- To generate the library, 8 individual electroporations are performed as follows.
- Take 50 μl of freshly-prepared electrocompetent YeO3-R1 cells. Mix on ice with 1.7 μl (990 ng; preferentially use 800-1,000 ng of complexes for highest efficiency) of Cat-Mu transposition complex preparation.
- Transfer the mixture to a pre-chilled 0.1-cm electrode spacing cuvette, and carry out electroporation using Genepulser II with the following settings: 200 Ω, 1.8 kV, 25 μF.
- Add 1 ml of pre-warmed SOC medium, and combine the suspensions from the individual electroporations in a 50-ml flask.
- Grow bacteria for 50 min at 28 °C with agitation (220 rpm).
- Add 3.3 ml 50% glycerol to the final concentration of 15% (v/v).
- Freeze the library as aliquots (e.g. 14 x 0.8-ml) in liquid nitrogen, and store at -80 °C.
- To estimate the number of individual transposon insertion mutants in the library thaw one aliquot and spread on LA plates with Chloramphenicol (10 μg/ml) (LA-Clm).
- Incubate 1–2 days at 28 °C (i.e., optimal growth temperature for Y. enterocolitica), and calculate the number of colony forming units (CFU).
- Isolation of enterocoliticin resistant mutants (Pinta et al., 2012)
- From the library aliquots prepared above spread a total of 16,000 CFU on LA-Clm plates such that you obtain individual colonies. Typically spread four library aliquots as 50 μl aliquots to 40-50 LA-Clm plates.
- Incubate 2 days at RT.
- Pool the Clm-resistant colonies by adding 1 ml PBS / plate and detach the colonies gently by scraping with an L-spreader.
- With a pipette transfer the PBS-suspension into a 50 ml Falcon tube.
- Pool the colonies from all other plates and combine them into one 50 ml tube. Mix the tube gently but thoroughly f. ex. by pulse vortexing.
- To prepare the pooled transposon library for long-time storage (i.e., the part you don't use immediately) follow steps 8-10.
- To continue with the isolation of enterocoliticin resistant mutants follow the steps from step 11 forward.
- Centrifuge the 50 ml tube at 3,000 rpm (2,000 x g) for 15 min at 4 °C and carefully remove most of the supernatant.
- Resuspend the pellet in 5 ml of 15% glycerol in TSB.
- Freeze the library as 0.5 ml aliquots and store at -80 °C.
- Measure the OD600 of the pooled transposon library from a suitable dilution (f.ex. 1:100).
- Based on the result dilute the cell suspension with PBS to OD600 ~0.2.
- Mix 10 μl of the OD600 ~0.2 suspension with 10 μl of enterocoliticin (Strauch et al., 2001) (suitable enterocoliticin-dose is determined in section VII) and 80 μl of TSB.
- Spread the mixture on a LA-Clm plate.
- Repeat steps 13-14 four times to get altogether 5 plates.
- Incubate the plates 1-2 days at RT to allow the growth of enterocoliticin-resistant mutants.
- Pick up single colonies from the plates and spread as pure cultures on new LA-Clm plates. These will constitute the candidate enterocoliticin-resistant mutants.
- Incubate the plates 1-2 days at RT.
- The enterocoliticin-resistance of the pure cultures has to be confirmed using a drop-test. Divide an LA-Clm plate into 8-12 sectors and culture the mutants on the sectors as a lawn using inoculation loops. As control, culture known enterocoliticin-sensitive and –resistant bacteria on some sectors.
- Immediately, apply a 2 μl drop of enterocoliticin in the center of each lawn.
- Incubate the plate 1-2 days at RT and inspect the sectors.
- he enterocoliticin-sensitive bacteria have no growth on the spot where enterocoliticin was applied while the enterocoliticin-resistant bacteria grow all over the sector.
Continue with the enterocoliticin-resistant mutants to section 8.
- Determination of the killing potential of the enterocoliticin solution
- Use a fresh colony of YeO3-R1 to inoculate 5 ml of TSB in a 15 ml tube. Grow the cells overnight with aeration at RT.
- Measure the OD600 of the culture. Dilute the culture in PBS so to OD600 ~0.2.
- Mix 10 μl of the OD600 ~0.2 suspension with different amounts of the enterocoliticin solution [Strauch et al., 2001] (f. ex. 1, 5, 10, 30 and 50 μl). Fill up the volume of the mixtures to 100 μl with TSB.
- Plate the different mixtures on separate LA-Clm plates.
- Incubate the plates 1-2 days at RT.
- Count the spontaneous resistant colonies. There should be less than ten spontaneous resistant colonies / plate.
- Identification of the transposon insertion site
The Cat-Mu transposon insertion sites can be identified by sequencing the transposon-containing genomic fragment that are cloned into pUC19 (Pinta et al., 2012).
- For the cloning of the transposon insertion fragment, HindIII digested genomic DNA (isolated using a DNA-isolation kit suitable for bacteria) of the mutant is cloned into HindIII digested pUC19 using standard protocols. HindIII does not cleave the Cat-Mu, thus ClmR transformants can be selected.
- The inserts in the plasmids isolated from the transformants can be sequenced using transposon-specific primers Muc1 and Muc2 (5'-GCTCTCCCCGTGGAGGTAAT-3' and 5'-TTCCGTCACAGGTATTTATTCGGT-3', respectively). The sequences obtained from these allow identification of the Cat-Mu -flanking regions on both sides of the transposon-insertions.
Recipes
- TGD buffer
10 mM Tris-HCl (pH 6.0)
0.5% glycerol
0.1 mM DTT - 5x complex buffer (10 ml)
3,750 μl 2 M Tris-HCl (pH 6)
125 μl 10% Triton X-100
2,500 μl 3 M NaCl
10 μl 0.5 M EDTA (pH 8)
3,615 μl ddH2O
Filter-sterilize (0.2 μm) and divide into aliquots.
Can be stored at -20 °C for 1 year - 1 M DTT (20 ml)
3.09 g DTT
Add 0.01 M Sodium acetate (NaAc) pH 5.2 to final volume.
Filter-sterilize (0.2 μm) and divide into aliquots.
Can be stored at -20 °C for 1 year. - 20% PEG6000 (w/v) - 2.5 M NaCl (5 ml)
1 g PEG6000
2.5 ml 5 M NaCl
Add ddH2O to final volume.
Filter-sterilize (0.2 μm) and use during the same day - SOB medium (1 L) without magnesium
20 g Bacto-tryptone
5 g Bacto-yeast extract
0.584 g NaCl
0.186 g KCl
Adjust pH to 7.0 with 5 M NaOH
Add ddH2O to final volume and autoclave
Stored at RT for up to 1 month - 2 M Mg++ stock
20.33 g MgCl2 · 6H2O
24.65 g MgSO4 · 7H2O
Add ddH2O to final volume.
Filter-sterilize (0.2 μm) or autoclave.
Stored at 4 °C for up to 6 months - 2 M glucose (100 ml)
26.04 g glucose
Add ddH2O to final volume
Filter-sterilize (0.2 μm)
Stored at 4 °C for up to 6 months - SOC medium (100 ml)
1 ml 2 M Mg++ stock
1 ml 2 M glucose
98 ml SOB medium without magnesium - Luria broth (LB) medium (1 L)
10 g Bacto-tryptone
5 g Bacto-yeast extract
5 g NaCl
Adjust pH to 7.0 with 5 M NaOH
Add ddH2O to final volume and autoclave
Stored at RT for up to 1 month - LB agar (LA) plates (1 L)
1 L LB medium
15 g Bacto-agar
Add Bacto-agar to LB medium before autoclaving
Cool down to 55 °C
Supplement with suitable antibiotics and dispense approx. 20 ml per Petri dish
Stored at 4 °C for up to 1 month - Chloramphenicol (Clm) solution (10 ml)
0.2 g Chloramphenicol
Add absolute ethanol to final volume
Stock solution (20 mg/ml) can be stored at –20 °C for 1 year
Chloramphenicol is used at the final concentration of 10 μg/ml - Tryptic soya broth
Prepare as instructed by the manufacturer - Phosphate-buffered saline (PBS) (pH 7.4) (1 L)
Prepare 1 L of 1x PBS as follows. Add to 800 ml of ddH2O 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4. Adjust the pH to 7.4 with HCl. Add distilled water to a total volume of 1 L. Dispense the solution into aliquots and sterilize by autoclaving (20 min, 121 °C, liquid cycle). Stored at RT. - TAE buffer
2 M Tris-acetate (pH 8.0)
50 mM EDTA
Acknowledgments
The protocol was adapted from our previously published paper Pinta et al. (2012). This work was supported by the Academy of Finland (projects 114075, 104361 and 50441) to M.S. and Finnish Glycoscience Graduate School to E.P. We thank Dr Eckhard Strauch for providing the enterocoliticin used in the work.
References
- al-Hendy, A., Toivanen, P. and Skurnik, M. (1992). Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect Immun 60(3): 870-875.
- Haapa, S., Taira, S., Heikkinen, E. and Savilahti, H. (1999). An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res 27(13): 2777-2784.
- Lamberg, A., Nieminen, S., Qiao, M. and Savilahti, H. (2002). Efficient insertion mutagenesis strategy for bacterial genomes involving electroporation of in vitro-assembled DNA transposition complexes of bacteriophage mu. Appl Environ Microbiol 68(2): 705-712.
- Pajunen, M. I., Pulliainen, A. T., Finne, J. and Savilahti, H. (2005). Generation of transposon insertion mutant libraries for Gram-positive bacteria by electroporation of phage Mu DNA transposition complexes. Microbiology 151(Pt 4): 1209-1218.
- Paatero, A. O., Turakainen, H., Happonen, L. J., Olsson, C., Palomaki, T., Pajunen, M. I., Meng, X., Otonkoski, T., Tuuri, T., Berry, C., Malani, N., Frilander, M. J., Bushman, F. D. and Savilahti, H. (2008). Bacteriophage Mu integration in yeast and mammalian genomes. Nucleic Acids Res 36(22): e148.
- Pinta, E., Li, Z., Batzilla, J., Pajunen, M., Kasanen, T., Rabsztyn, K., Rakin, A. and Skurnik, M. (2012). Identification of three oligo-/polysaccharide-specific ligases in Yersinia enterocolitica. Mol Microbiol 83(1): 125-136.
- Strauch, E., Kaspar, H., Schaudinn, C., Dersch, P., Madela, K., Gewinner, C., Hertwig, S., Wecke, J. and Appel, B. (2001). Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl Environ Microbiol 67(12): 5634-5642.
Article Information
Copyright
© 2012 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pajunen, M., Pinta, E. and Skurnik, M. (2012). Construction and Screening of a Transposon Insertion Library of Yersinia enterocolitica (YeO3-R1). Bio-protocol 2(15): e246. DOI: 10.21769/BioProtoc.246.
Category
Microbiology > Microbial genetics > Mutagenesis
Molecular Biology > DNA > Mutagenesis
Systems Biology > Genomics > Transposons
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










