- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Guard-cell Enriched Tissue for RNA Extraction
Published: Vol 7, Iss 15, Aug 5, 2017 DOI: 10.21769/BioProtoc.2447 Views: 16035
Reviewed by: Scott A M McAdamYi ZhangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurements of Root Colonized Bacteria Species
Vílchez Juan Ignacio [...] Huiming Zhang
Apr 5, 2021 6321 Views

Transgene-free Genome Editing in Grapevine
Edoardo Bertini [...] Sara Zenoni
Feb 20, 2025 2505 Views

Isolation and Transfection of Protoplasts From Maize Mesophyll Cells
Lauren A. Higa [...] Zhi-Yan Du
Feb 5, 2026 64 Views
Abstract
This is a protocol for isolation of guard cell enriched samples from Arabidopsis thaliana plants for RNA extraction. Leaves are blended in ice-water and filtered through nylon mesh to obtain guard cell enriched fragments. With guard cell enriched samples, gene expression analysis can be done, e.g., comparing different gene expression levels in guard cells versus whole leaf to determine if a gene of interest is predominantly expressed in guard cells. It can also be used to study the effect of treatments or different genetic backgrounds in the regulation of the guard cell expressed genes.
Keywords: Arabidopsis thalianaBackground
Isolation of guard cells for RNA extraction has traditionally relied on guard cell protoplast extraction (Leonhardt et al., 2004) or epidermal peels (Pandey et al., 2010). Manual dissection of guard cells from freeze-dried leaves has also been used (Bates et al., 2012). The protoplast procedure may introduce unwanted changes in gene expression from wounding effects and other methods are time consuming, require special equipment or expensive reagents (e.g., transcriptional inhibitors). Hence there is a need for a fast and simple protocol to isolate guard cells for gene expression analysis. Here we further describe a quick ice blender method for isolation of guard cell enriched tissue (Bauer et al., 2013), in the form of epidermal fragments without mesophyll and other vascular cells. This method uses a nylon mesh to collect epidermis from blended leaf tissue. The adequately large pore size of the nylon mesh allows mesophyll and other vascular cells to pass through while retaining the epidermal fragments. We propose that the critical factor in this protocol is the type of blender used to process the samples.
Materials and Reagents
- Razor blade
- 210 μm nylon mesh (A. Hartenstein, laborversand.de, catalog number: PAS1 )
- Plastic embroidery hoop from a handicraft store (Figure 1)
Note: Nylon mesh is placed in the plastic embroidery hoop to provide a little tension to the mesh, to make it easier to pour the blended water and guard cell enriched fragments through the mesh.
Figure 1. Nylon mesh with a plastic embroidery hoop - Paper towel
- Small medical spatula or plastic spoon
- 1.5 ml tube or aluminum foil
- Arabidopsis plants
- Milli-Q or distilled water kept at 4 °C
- Crushed ice
- Liquid nitrogen
Equipment
- Blender (Braun JB 3060 or other 800 W blender) (Braun Household, model: JB 3060 )
Note: A blender with lower effect has been tried, but failed to isolate proper fragments, probably due to insufficient power to fully crush the ice. - Microscope (ZEISS, model: SteREO Discovery.v20 )
Procedure
Collect leaves from 4-7 week old Arabidopsis plants grown in soil at 12 h/12 h photoperiod, use 17-18 plants and 4-5 leaves per plant. If plants are smaller in size, use more leaves per plant.
- Remove the central vein (midrib) by razor blade. Collect about 1.0-1.5 g of leaves without midribs for the following procedures (Figure 2).

Figure 2. Midrib is cut and removed with the razor blade - Put the cut leaf blades in blender with 250 ml cold Milli-Q water and a handful of crushed ice.
Blend 1 min (program 2 for the Braun blender), pour through mesh placed in a plastic embroidery hoop.
Note: Pouring through mesh should be done slowly, so that the guard cell enriched fragments are collected in the middle of the mesh. This is especially important at step 7 after which the sample is collected. - Wash the mesh with cold Milli-Q water above the blender cup so that the guard cell enriched fragments are transferred inside the blender cup. Add cold Milli-Q water up to 250 ml and handful of crushed ice.
- Blend 1 min (program 2), pour through mesh placed in a plastic embroidery hoop.
- Wash the mesh with cold Milli-Q water above the blender cup so that the guard cell enriched fragments are transferred inside the blender cup. Add cold Milli-Q water up to 250 ml and handful of crushed ice.
- Blend 1 min (program 2), pour through mesh placed in a plastic embroidery hoop.
- Remove the remaining dark green tissue fragments (small leaf pieces that have not been blended) from the light green epidermal fraction.
- Dry the underside of the mesh from excess water with paper towel about 10-15 sec and use a small spatula or plastic spoon to carefully scrape the sample into a 1.5 ml tube or aluminum foil (Figure 3).

Figure 3. Guard cell enriched sample that is collected into a 1.5 ml tube - Freeze the sample in liquid nitrogen and keep at -80 °C. Alternatively, the sample can be immediately ground in mortar with liquid nitrogen and the powder used for RNA isolation.
Note: If an estimation of guard cell enrichment is required, one easy control is to harvest a whole leaf sample from the same plants used for guard cell enrichment and freeze in liquid nitrogen and store at -80 °C. RNA can then be extracted in parallel with the guard cell samples. The level of guard cell enrichment can be estimated by real time quantitative PCR using guard cell preferentially expressed genes e.g., HT1 (AT1G62400), OST1 (AT4G33950) or GORK (AT5G37500).
Data analysis
Isolated guard cell enriched fragments can be visualized with a stereo microscope (Zeiss SteREO Discovery.v20), and no mesophyll fragments (green tissue) should be present (Figure 4A). While testing different blenders, successful isolation of guard cell enriched fragments was achieved with the Braun (800 W) blender but not with a blender with lower effect (Figure 4).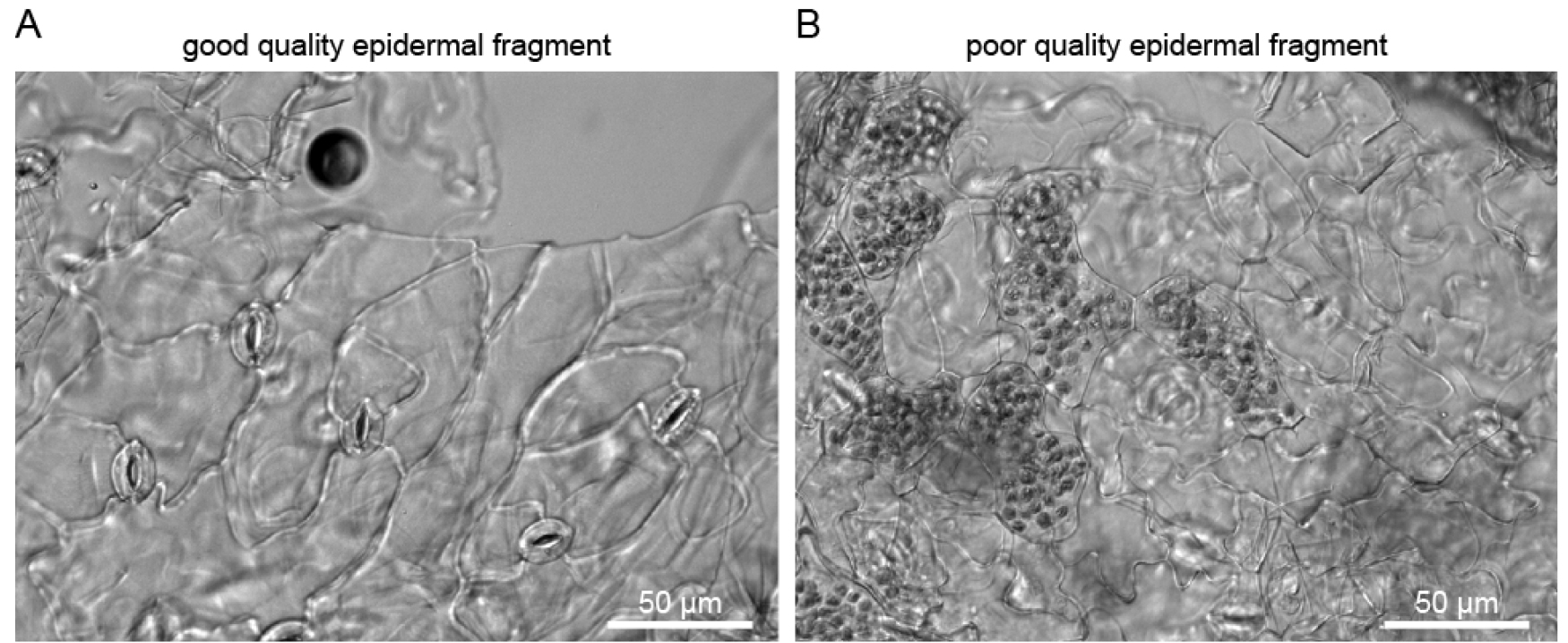
Figure 4. Isolated guard cell enriched fragments under a microscope. A. Sample isolated with the Braun blender; B. Sample isolated with a small 220 W blender.
To control the quality of guard cell enrichment, real-time quantitative PCR can be done with guard cell expressed genes like HT1 and GORK, by comparing their expression level in guard cell enriched sample versus a whole leaf sample (Figure 5; for details about the qPCR conditions see Jalakas et al., 2017).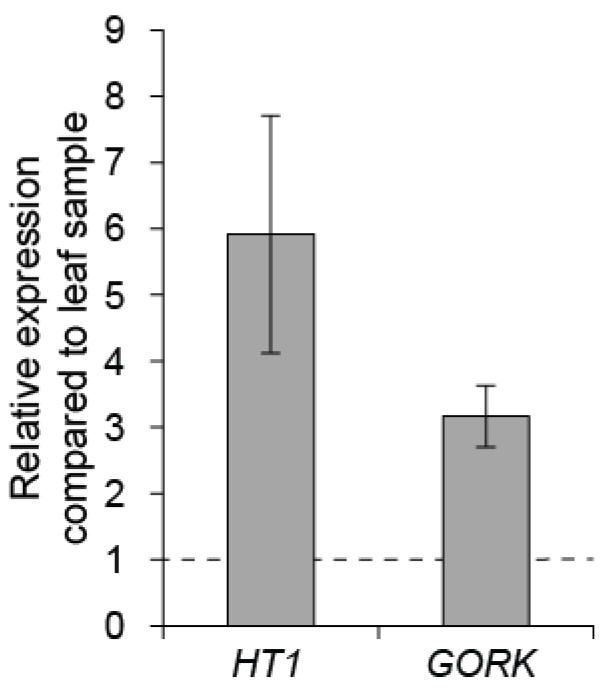
Figure 5. Gene expression in Col-0 guard cell enriched sample compared to leaf sample. The mean of three biological replicates are shown; error bars depict ± SEM.
Acknowledgments
This protocol was adapted from the research article Bauer et al. (2013). This work was funded by the Estonian Ministry of Science and Education (IUT2-21 to H.K.), the European Regional Development Fund (Center of Excellence in Molecular Cell Engineering CEMCE to H.K.), the Academy of Finland (grant number #307335, Center of Excellence in Primary Producers 2014-2019 to M.B.).
References
- Bates, G. W., Rosenthal, D. M., Sun, J., Chattopadhyay, M., Peffer, E., Yang, J., Ort, D. R. and Jones, A. M. (2012). A comparative study of the Arabidopsis thaliana guard-cell transcriptome and its modulation by sucrose. PLoS One 7(11): e49641.
- Bauer, H., Ache, P., Lautner, S., Fromm, J., Hartung, W., Al-Rasheid, K. A., Sonnewald, S., Sonnewald, U., Kneitz, S., Lachmann, N., Mendel, R. R., Bittner, F., Hetherington, A. M. and Hedrich, R. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol 23(1): 53-57.
- Jalakas, P., Huang, Y.-C., Yeh, Y.-H., Zimmerli, L., Merilo, E., Kollist, H. and Brosché, M. (2017). The role of ENHANCED RESPONSES TO ABA1 (ERA1) in Arabidopsis stomatal responses is beyond ABA signaling. Plant Physiol 174: 665-671.
- Leonhardt, N., Kwak, J. M., Robert, N., Waner, D., Leonhardt, G. and Schroeder, J. I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16(3): 596-615.
- Pandey, S., Wang, R. S., Wilson, L., Li, S., Zhao, Z., Gookin, T. E., Assmann, S. M. and Albert, R. (2010). Boolean modeling of transcriptome data reveals novel modes of heterotrimeric G-protein action. Mol Syst Biol 6: 372.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Jalakas, P., Yarmolinsky, D., Kollist, H. and Brosche, M. (2017). Isolation of Guard-cell Enriched Tissue for RNA Extraction. Bio-protocol 7(15): e2447. DOI: 10.21769/BioProtoc.2447.
- Jalakas, P., Huang, Y.-C., Yeh, Y.-H., Zimmerli, L., Merilo, E., Kollist, H. and Brosché, M. (2017). The role of ENHANCED RESPONSES TO ABA1 (ERA1) in Arabidopsis stomatal responses is beyond ABA signaling. Plant Physiol 174: 665-671.
Category
Plant Science > Plant cell biology > Cell isolation
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link