- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bioelectrospray Methodology for Dissection of the Host-pathogen Interaction in Human Tuberculosis
Published: Vol 7, Iss 14, Jul 20, 2017 DOI: 10.21769/BioProtoc.2418 Views: 9927
Reviewed by: Alka MehraMarc-Antoine SaniAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views

A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1251 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 92 Views
Abstract
Standard cell culture models have been used to investigate disease pathology and to test new therapies for over fifty years. However, these model systems have often failed to mimic the changes occurring within three-dimensional (3-D) space where pathology occurs in vivo. To truthfully represent this, an emerging paradigm in biology is the importance of modelling disease in a physiologically relevant 3-D environment. One of the approaches for 3-D cell culture is bioelectrospray technology. This technique uses an alginate-based 3-D environment as an inert backbone within which mammalian cells and extracellular matrix can be incorporated. These alginate-based matrices produce highly reproducible results and can be mixed with different extracellular matrix components. This protocol describes a 3-D system incorporating mycobacteria, primary human blood mononuclear cells and collagen-alginate matrix to dissect the host-pathogen interaction in tuberculosis.
Keywords: BioelectrosprayBackground
Mycobacterium tuberculosis (Mtb) is a pathogen of global public health importance that causes a mortality of 1.8 million people per year and morbidity of 10 million worldwide (WHO, 2016). Despite substantial investment in research, much greater understanding of the host-pathogen interaction is required to improve prevention and treatment. Currently, the pathogen is becoming increasingly resistant to commonly used drugs, with the emergence of extensively drug-resistant Mtb. One of challenges to the tuberculosis (TB) field is the availability of model systems to interrogate the host-pathogen interaction, as widely used animal models do not fully reflect pathology in humans. Hence, there is an urgent need to complement these animal models by developing a physiologically relevant in vitro environment (Bielecka et al., 2017; Tezera et al., 2017). Mtb is an obligate pathogen of man and so we hypothesized that a model system requires human cells, virulent mycobacteria, 3-dimensional organization, extracellular matrix, longitudinal readouts and the ability to modulate the environment over time.
This protocol describes a physiologically relevant in vitro environment by utilizing human cells, extracellular matrix components and live Mtb using a bioelectrospray model to mimic human Mtb infection. This 3-D model is different from other models by using an extracellular matrix that can be released by de-capsulation so that downstream analysis of cells within the matrix can be performed. Furthermore, this methodology has wide potential applicability to investigate infectious, inflammatory and neoplastic diseases and develop novel drug regimens and vaccination approaches.
Materials and Reagents
- Female Luer Thread Style Connectors (West Group, catalog number: FTLL210-J1A )
- Tissue paper
Note: Sterilize lab tissue paper by autoclaving and keep it sterile until use. - Nozzle for bioelectrosprayer (0.7 φ) (2 pieces) (Nisco Engineering, catalog number: PE-00577 )
- Silicon tubes with connector attached at the end (3 pieces) (VWR, catalog number: 228-0705 )
- FalconTM 50 ml conical centrifuge tubes (Corning, Falcon®, catalog number: 352070 )
- 75 cm2 flask (CELLSTAR® Cell Culture Flasks, Greiner Bio One International)
- SterilinTM 7 ml polystyrene Bijou containers (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 129A )
- 2 ml Eppendorf tubes
- 20 ml syringe
- 5 ml syringe
- 0.22 µm syringe filter EMD Millipore MillexTM-GP 33mm (Fisher Scientific, catalog number: 10268401)
Manufacturer: EMD Millipore, catalog number: SLGP033RS . - Pipette (10 ml, 25 ml) (CELLSTAR® Serological Pipettes , Greiner Bio One International)
- Mycobacterium tuberculosis
Note: Any strain or different species can be used. - Methylated spirit (Fisher Scientific, catalog number: 11492874 )
- RPMI 1640 medium (Thermo Fisher Scientific, GibcoTM, catalog number: A1049101 )
- EDTA powder (Fisher Scientific, catalog number: 10522965 )
- Versene solution (Thermo Fisher Scientific, GibcoTM, catalog number: 15040033 )
- Hank’s balanced salt solution (HBSS), no calcium, no magnesium (Thermo Fisher Scientific, GibcoTM, catalog number: 14170112 )
- HBSS, calcium, magnesium (Thermo Fisher Scientific, GibcoTM, catalog number: 24020117 )
- Human AB serum (Sigma-Aldrich, catalog number: H4522-100ML )
- Water (Millipore double distilled water, sterile)
- Ultrapure alginate PRONOVATM UP MVG 10 mg (FMC, NovaMatrix, catalog number: 4200106 )
- 1 N NaOH cell culture grade solution (Sigma-Aldrich, catalog number: S2770-100ML )
- 1 M HEPES (Thermo Fisher Scientific, GibcoTM, catalog number: 15630106 )
- 7.5% NaHCO3 solution (Thermo Fisher Scientific, catalog number: 25080060 )
- Human collagen type I (3 mg/ml) (Advanced Biomatrix, catalog number: 5007-A )
- 0.01 N HCl (pH = 2.0)
- Calcium chloride, 96%, extra pure, powder, anhydrous, ACROS OrganicsTM (Thermo Fisher Scientific, catalog number: 10021681)
Manufacturer: Acros Organics, catalog number: 349615000 . - Sodium citrate dihydrate (Fisher Scientific, catalog number: 10396430 )
- 3% alginate (w/v) (see Recipes)
- Collagen-alginate mix (1:1 ratio) (see Recipes)
- CaCl2 precipitation bath (see Recipes)
- De-capsulating solution (see Recipes)
Equipment
- Oven (Genlab Drying Oven, UK)
- Plastic beakers (2.5 L, Thermo Fisher Scientific)
- Class I/III biosafety cabinet
- Borosilicate crystalizing glass beakers with spout (VWR, catalog number: 216-0068 ) (5 beakers)
- Magnetic stirrers (1 cm, 5 pieces)
- Sterile scissors
- Thermo ScientificTM NalgeneTM Polypropylene Scissor-Type forceps (Thermo Fisher Scientific, Thermo ScientificTM)
- 37 °C, 5% CO2 incubator
- Centrifuge (Eppendorf, model: 5427 R )
- Electrostatic Bead Generator VARv1 (Nisco Engineering, model: VAR v1 )
- Test-Tube-rotator (Bibby Scientific, model: STR4 )
- PHD ULTRATM CP syringe pump (Harvard Apparatus, model: PHD ULTRATM CP Syringe Pump )
- Jack for height adjustment (Nisco Engineering, catalog number: PE-01162 )
Procedure
- SOP for bioelectrospray pre-infection
Note: This is the optimized protocol for encapsulation of peripheral blood mononuclear cells (PBMCs) after overnight infection with Mycobacterium tuberculosis. If the organism of interest is Mycobacterium tuberculosis, the reader should assume that the experiment is done under Class I/III biosafety cabinet in a standard biosafety level 3 lab under approved institutional standards of practice. If the work requires being undertaken in a standard biosafety level 2 lab, one can fumigate the whole machine according to the respective laboratory SOP before taking out from the BSL3 laboratory. The bioelectrospray technique is always performed in a Class I/III biosafety cabinet, with the doors of the bioelectrosprayer kept closed during microsphere generation as an extra level of containment. A Class II biosafety cabinet is inherently not safe to do the procedure. Once the microspheres are formed, they can be transferred for the subsequent steps either to Class II biosafety cabinet or continue in Class I/III biosafety cabinet. The protocol spans three days. Modification for other infections or other biological modelling is possible.
Day-1- Sterilize the following items in plastic beakers and dry them in an oven.
- Five 150 ml Borosilicate glass beakers with spout
- Magnetic stirrers (1 cm length) (5 pieces)
- Female Luer Thread Style Connectors
- Scissors
- Forceps (different sizes)
- Tissue paper
- Sterile silicon tubes with connector attached at the end (3 pieces, 45 cm in length)
- Prepare alginate suspension. Alginate is a natural product and there is variability on the product depending on the species of alginate and environmental conditions. In all studies our group have conducted so far, all the procedures were done with medium viscosity G dominant alginate with viscosity above 250 mPas.
- Prepare 3% alginate in HBSS without Ca/Mg, with phenol red under sterile conditions and mix it with the buffers to form alginate mix.
Day-2- Prepare the alginate-collagen matrix as in Recipe 2 (Figure 1).
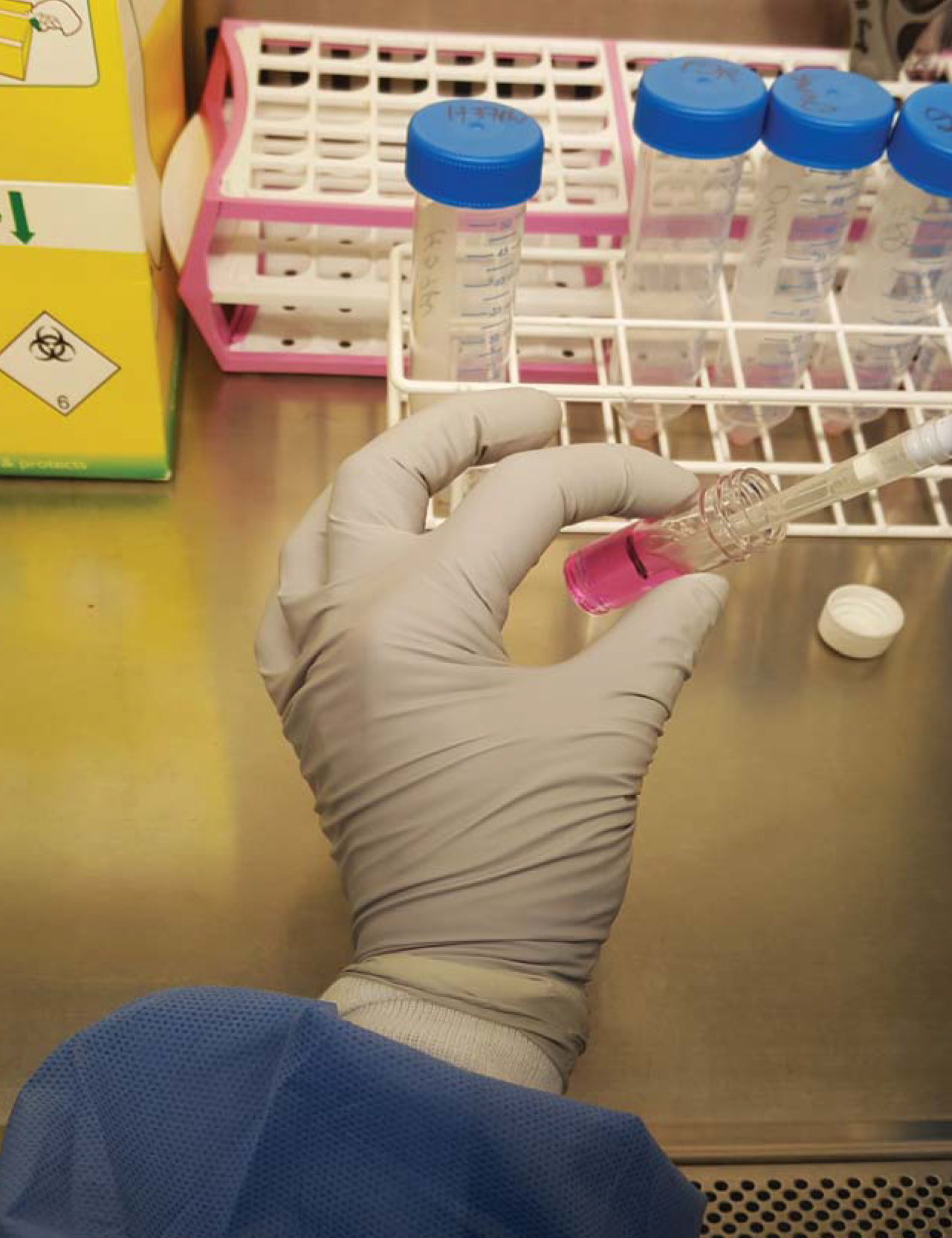
Figure 1. Preparation of cells for encapsulation. Cells are recovered from a 75 cm2 flask and pelleted in a Falcon by centrifugation, and then mixed with alginate-collagen matrix in a 7 ml bijou container (usually 25 million cells/5ml of matrix mix). Also see Video 1.Video 1. Mixing alginate with PBMCs prior to bioelectrospraying - Extract PBMCs according to standard protocol for separating PBMCs from blood by density centrifugation. One can use either the buffy coat cells which are commercially available or whole blood isolated PBMCs.
- Before the final wash, re-suspend the cells in 50 ml of HBSS without Ca/Mg and take 15 µl of cell suspension for cell counting (dilute 10x further if working with leukocyte cones isolated from 500 ml blood). Place the re-suspended cells in the fridge until use.
- Count the cells, and calculate the total cells required for final concentration of 5 x 106 cells/ml once re-suspended in cell-alginate-mix.
- Pipette off appropriate number of isolated PBMCs and then pellet by spinning at 320 x g, 8 min, 4 °C, in a 50 ml Falcon tube. Discard supernatant and add 30 ml of complete RPMI medium (ampicillin, glutamine) to the PBMC pellet and re-suspend.
- Infect PBMCs with appropriate multiplicity of infection (MOI) of Mtb (our experimental standard is MOI 0.1).
- Transfer the infected PBMCs into a 75 cm2 flask.
- Leave overnight in a 37 °C, 5% CO2 incubator.
Day-3- Preparation of cells for encapsulation
- Next day, take out the flask from the incubator; carefully transfer the contents of the flask (30 ml) to a 50 ml Falcon tube.
- Add 5 ml of 5 mM EDTA (or Versene, Thermo Fisher Scientific) to the flask and incubate for 8-10 min at 37 °C, 5% CO2 incubator.
- After that time, add 5 ml of HBSS without Ca, Mg (or complete RPMI medium) to the flask to dilute the effect of the detachment solution.
- Scrape the bottom surface of the flask with a scraper carefully and lightly to re-suspend all remaining cells.
- Transfer the 10 ml contents to the same 50 ml Falcon tube, already containing the medium (total of 40 ml). Finally rinse flask with 10 ml of HBSS without Ca, Mg (or complete RPMI medium) and add to the Falcon.
- Pellet cells in the Falcon by centrifugation at 320 x g, 8 min.
- Carefully, take the Falcon tube out of centrifuge and decant supernatant. Re-suspend the pelleted cells: e.g., 50 µl per 5 x 106 cells.
- Prepare 5 ml alginate-collagen mix in a 7 ml bijou container. You can prepare alginate-collagen mix up to a week prior to the experiment (Video 1).
- Mix well your cells with the alginate-collagen mix accordingly in a 7 ml bijou container (Usually 25 million cells/5 ml of alginate).
- Keep at 4 °C on ice/in the fridge until bioelectrospraying the cell-alginate suspension.
- Next day, take out the flask from the incubator; carefully transfer the contents of the flask (30 ml) to a 50 ml Falcon tube.
- Bioelectrospray (Figures 2, 3 and 4)

Figure 2. Nisco electrostatic encapsulator with washed and alcohol sterilized arm, sterile nozzle, sterile silicon tubes and crystalizing glass beakers. 1. Nozzle (0.7 φ) attached to the nozzle holder; 2. Electrostatic accelerator arm for the electrostatic bead generator VARv1; 3. Electrode cable; 4. Silicon tubes with connector attached at the end; 5. Borosilicate crystalizing glass beakers with spout with magnetic stirrers (1 cm); 6. Stirrer; 7. Ring on the electrostatic accelerator arm; 8. Ruler for setting correct needle height.
Figure 3. Biolelectrospraying microspheres. Matrix in syringe is injected to the bioelectrosprying machine and microspheres are formed. A syringe filled with matrix was set up on syringe pump (A) so that it will inject the matrix into silicon tube (B) connected to the electrostatic bead generator. The syringe driver is sitting on jack (C) for height adjustment. E. Unused crystalizing glass beakers on the roof of the bioelectrospray machine; F. Housing with doors to enclose the electrostatic bead generator; G. High-voltage switch on/off (white, on/green, off) which is left of potentiometers for optional peristaltic pump, agitator speed and voltage on electrode. Voltage indicator displaying 7.0 kV. Biobin (H) and old media bottle (I) containing surfanios (10%) for discarding biohazardous waste.
Figure 4. Microspheres are formed in HBSS solution with 100 mM CaCl2. A mix of collagen-alginate with cells in 2 mm diameter silicon tube at a specific rate and microspheres are formed in the gelling bath. Also see Video 3. - Items required:
- Bioelectrospray sterile items (Listed above Procedure A Day-1)
- Nisco Electrostatic Encapsulator with washed and alcohol sterilized arm
- 50 ml Falcon tube
- 1 M CaCl2 solution
- HBSS with Ca, Mg
- HBSS without Ca/Mg
- Bioelectrospray sterile items (Listed above Procedure A Day-1)
- Procedure:
- Set the machine up at the rear of the MSC class I/III with the syringe driver on the jack adjacent to it, so that the syringe driver is equal height to the top of the bioelectrosprayer (see Videos 2 and 3).Video 2. Setting up the bioelectrospray systemVideo 3. Bioelectrospray system in operation with microspheres being formed
- Adjust the syringe driver speed according to the diameter of the syringe (e.g., appropriate rate for 5 ml syringe to give 10 ml/h). The adjustment of the speed varies dependent on the syringe brand.
- Prepare the Borosilicate glass beaker by placing magnetic stirrer inside.
- Open the sterile silicon tubes and the nozzles and connect them to the bioelectrospray needle held in the arm of encapsulator.
- Run 50 ml HBSS without Ca/Mg through the tubing using a 20 ml syringe slowly into empty Borosilicate glass beaker. This will wash the system and check the connection of the needle in the encapsulator arm.
- Dry the arm with sterile tissue.
- Aspirate the cell-matrix mixture into a 5 ml syringe slowly. Alginate is very viscous and so this must be performed with patience. Avoid creating bubbles. The air enclosed will be ultimately end inside the microspheres, causing them to float.
- Connect the 5 ml syringe to the tube, and inject it slowly until it nears the bioelectrospray needle at the end.
- Pour 100 mM CaCl2 in HBSS (without Ca/Mg) into one of the beakers until it is half-full. Put beaker under the arm of encapsulator.
- Place the syringe in the syringe driver and ensure driving screw abuts end of syringe. Close the doors of the bioelectrosprayer fully.
- Start bioelectrospraying by turning on the voltage and stirrer of the encapsulator, and initiating the syringe pump at 10 ml/h. Alginate-collagen mix will be ejected through the needle and the spheroids will be collected in the gelling bath. We use 7 kV voltage and 70% stirring speed. The voltage, stirring speed, affects the diameter of the microspheres and alginate type and nozzle size and further information can be found in the work of Workman and colleagues (Workman et al., 2014).
Safety Note: The electrostatic bead generator has an electric charge. Therefore, do not touch any parts when the generator is on to prevent exposure to high voltage (low current) electricity. - Once the syringe contents have been fully expelled by syringe driver, it is necessary to drive alginate mix from tubing dead space through the bioelectrosprayer needle or continue with second batch of the cell-matrix suspension.
- Stop the bioelectrosprayer, replace the syringe with one containing 5 ml HBSS (without Ca/Mg), and recommence bioelectrosprayer and driver at 10 ml/h until all collagen-alginate mix is expelled and HBSS reaches the needle. This will be clear from colour and microspheres no longer form in gelling bath.
- Once all alginate is bioelectrosprayed, decant microspheres to 50 ml tubes by pouring. Allow them to settle and then remove as much supernatant CaCl2 solution from the Falcon as possible to the waste bottle with a 5 ml pipette. Microspheres take about 2 min to settle and so centrifugation is not required, and may damage the spheres (Figure 5).
- Add HBSS with Ca, Mg to the microspheres to total volume 50 ml. Stand Falcons in racks.
- Wash the microspheres 2 x by removing HBSS with Ca, Mg with a pipette and then adding again (Video 4).
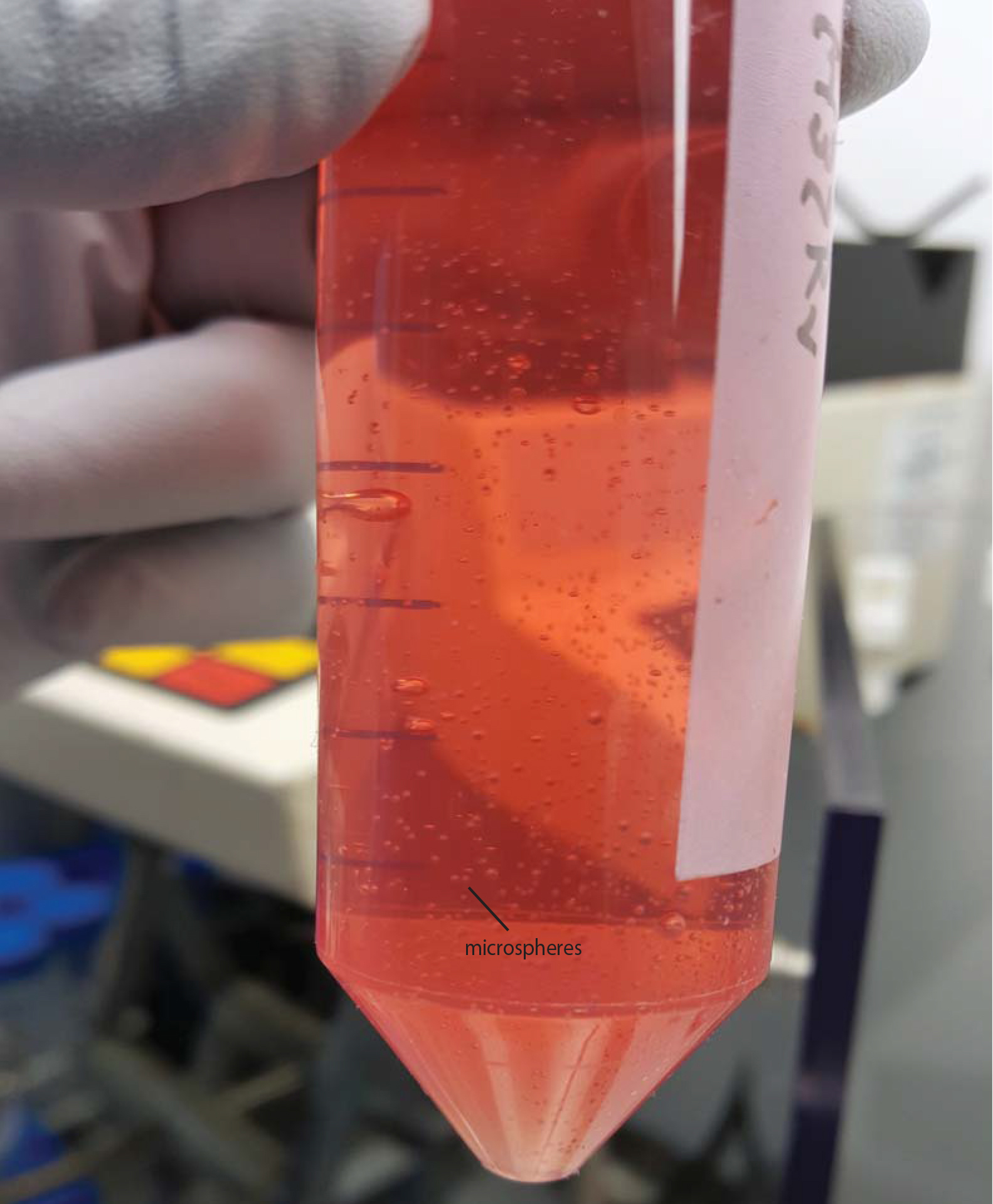
Figure 5. Microspheres in HBSS with Ca/Mg after transferred from the gelling bath. Also see Video 4.Video 4. Decanting microspheres after generation - Aliquot microspheres to the appropriate tissue culture plate or sterile Eppendorf tubes. Aliquoting is performed using a 1 ml pipette with the end cut off with sterile scissors, to give an orifice sufficiently large to pipette up microspheres. Keep the Falcon with microspheres agitated during pipetting to keep a constant concentration within the media and avoid settling during aliquoting.
- Pipette off HBSS from wells/Eppendorfs to leave microspheres only.
- Add RPMI 1640 medium supplemented with 10% AB serum, and incubate at 37 °C for the duration of the experiments. Each microsphere has ~600 µm diameter and the viability of the cells after a complete procedure is 95%.
- If setting up a second experimental condition (e.g., uninfected cells, different cell augmentation), then replace silicone tube and repeat steps as above. A maximum of 8 experimental conditions can be readily undertaken in one day. One should plan 1 h per 5 ml matrix, giving a total of up to 10 h for the final generation of microspheres.
- De-capsulation of cells
- Aspirate the microspheres into a 50 ml centrifuge tube and allow them to settle at the bottom of the tube. Then discard the supernatant carefully.
- Wash the microspheres with HBSS without Ca/Mg twice. Let the microspheres settle at the bottom of the tube, then remove the supernatant.
- Add 10 ml de-capsulating solution to the capsules.
- Mix the suspension thoroughly prior to incubation at room temperature up to 15 min. Shake the microspheres intermittently.
- When the microspheres are completely de-capsulated, their absence is visible with the naked eye. Fill the remaining tube with HBSS without Ca/Mg (or complete RPMI) at room temperature.
- Centrifuge the de-capsulated cells to pellet them at 320 x g for 5 min and discard the supernatant.
- Wash the cell pellet by re-suspending in HBSS followed by centrifugation.
- Further use of collagenase is usually not necessary.
- The de-capsulated cells can be further cultured as a monolayer or used for downstream analysis.
- Aspirate the microspheres into a 50 ml centrifuge tube and allow them to settle at the bottom of the tube. Then discard the supernatant carefully.
Data analysis
Once the microspheres are formed, one can consider them as an individual’s cells/collection of cells as the microspheres are permeable, like sponges. Microspheres can be set up in 96-well plates for assays testing viability, necrosis and apoptosis. Mycobacterial growth can be measured by luminescence if the bacterium has a luminescence reporter plasmid, or directly by counting colony forming units on Middlebrook 7H11 agar after de-capsulation. Cellular composition can be analyzed by flow cytometry after de-capsulation and paraformaldehyde fixation, and RNA analysis by de-capsulation and cellular lysis with Trizol. Data is analyzed according to standard workflows, our routine is a minimul of 2 separate donors with each experimental variable in triplicate.
Recipes
- 3% alginate (w/v)
- Measure the alginate in aseptic conditions in BSC-II
- Place approximately 1.5 g of purified sodium alginate (MVG from NovaMatrix with high glucuronic acid content ≥ 60%, viscosity > 200 mPas, and endotoxin ≤ 100 EU/g) in a sterile 50 ml tube
- Weigh sterile empty Falcon tube
- Weigh the Falcon tube with alginate
- Determine the weight of alginate by subtracting the weight of empty Falcon tube
- Add the appropriate volume of HBSS without Ca/Mg, for final percentage of 3%
- Vortex the tube for approximately 3 min to partially dissolve the alginate powder then place the tube on an orbital mixer at 10 x g overnight or for two nights in a cold room
- Store the alginate solution at 4 °C for short-term storage (1-2 weeks) or at -20 °C for long-term storage (one year)
- Collagen-alginate mix (1:1 ratio)
- Prepare the following buffers
- 0.05 N NaOH in 0.2 M HEPES by mixing 2.5 ml NaOH with 10 ml HEPES (stock 1 M) and adding 37.5 ml of endotoxin free water for final 50 ml solution of NaOH/HEPES solution
- 100 ml 7.5% NaHCO3
- Human collagen (3 mg/ml dissolved aqueous solution in 0.01 N HCl [pH = 2.0])
- Mix the HEPES/NaOH buffer, NaHCO3, and alginate as follows:
- 3% alginate: 50% of the total mix
- HEPES/NaOH: 4.5% of the total mix
- 7.5% NaHCO3: 9% of the total mix
- Filter sterilize by 0.22 µm filter (Fisher Scientific)
- Add the human collagen (3 mg/ml): 36.5% of the total mix
- Store the collagen-alginate solution at 4 °C for short-term storage (1-2 weeks) or at -20 °C for long-term storage (up to a year)
- CaCl2 precipitation bath
- Dissolve 147 g of CaCl2·2H2O and 23.8 g of HEPES in 1 L of Milli-Q H2O
- Adjust the pH level to 5-6
- Sterilize the solution using a 0.22 μm filter
- Precipitation fluid can be stored at room temperature (6-12 months)
- On the day of the experiment, mix 5 ml concentrate with 45 ml of HBSS without Ca/Mg for working solution
- De-capsulating solution
- Prepare and sterilize the following solutions
- 1 M of NaCitrate (add 294.1 g in 1 L of Milli-Q H2O)
- 1 M of EDTA (add 292.2 g in 1 L of Milli-Q H2O)
- 1 M of citric acid (add 192.1 g in 1 L of Milli-Q H2O)
- Store solutions in cell culture grade plastic containers at room temperature (6-12 months)
- Make 55 mM NaCitrate and 10 mM EDTA in HBSS with Ca, Mg and adjust the pH to 7.2
- To prepare 100 ml of de-capsulating solution
Mix 5.5 ml of NaCitrate and 1 ml of EDTA in HBSS with Ca, Mg and adjust the pH to 7.2-7.4 and store at 4 °C for up to 2 weeks
Acknowledgments
We would like to thank S. N. Jayasinghe from University College London, United Kingdom for all the technical support and advice on the bioelectrospray technology. This work is funded by the grant from the US National Institute for Health R33AI102239, the UK National Centre for the 3Rs NC/L001039/1 and the Antimicrobial Resistance Cross Council Initiative supported by the seven research councils MR/N006631/1.
References
- Al Shammari, B., Shiomi, T., Tezera, L., Bielecka, M. K., Workman, V., Sathyamoorthy, T., Mauri, F., Jayasinghe, S. N., Robertson, B. D., D'Armiento, J., Friedland, J. S. and Elkington, P. T. (2015). The extracellular matrix regulates granuloma necrosis in tuberculosis. J Infect Dis 212(3): 463-473.
- Bielecka, M. K., Tezera, L. B., Zmijan, R., Drobniewski, F., Zhang, X., Jayasinghe, S. and Elkington, P. (2017). A bioengineered three-dimensional cell culture platform integrated with microfluidics to address antimicrobial resistance in tuberculosis. mBio 8:e02073-16.
- Tezera, L. B., Bielecka, M. K., Chancellor, A., Reichmann, M. T., Shammari, B. A., Brace, P., Batty, A., Tocheva, A., Jogai, S., Marshall, B. G., Tebruegge, M., Jayasinghe, S. N., Mansour, S. and Elkington, P. T. (2017). Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. eLife 6:e21283.
- WHO. (2016). Global tuberculosis report 2016.
- Workman, V. L., Tezera, L. B., Elkington, P. T. and Jayasinghe, S. N. (2014). Controlled generation of microspheres incorporating extracellular matrix fibrils for three-dimensional cell culture. Adv Funct Mater 24(18): 2648-2657.
Article Information
Copyright
Tezera et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Tezera, L. B., Bielecka, M. K. and Elkington, P. T. (2017). Bioelectrospray Methodology for Dissection of the Host-pathogen Interaction in Human Tuberculosis. Bio-protocol 7(14): e2418. DOI: 10.21769/BioProtoc.2418.
- Tezera, L. B., Bielecka, M. K., Chancellor, A., Reichmann, M. T., Shammari, B. A., Brace, P., Batty, A., Tocheva, A., Jogai, S., Marshall, B. G., Tebruegge, M., Jayasinghe, S. N., Mansour, S. and Elkington, P. T. (2017). Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. eLife 6:e21283.
Category
Microbiology > Microbe-host interactions > Bacterium
Immunology > Immune cell function > Macrophage
Cell Biology > Cell isolation and culture > 3D cell culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









