- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of the Dot/Icm Type IV Secretion System Core Complex from Legionella pneumophila for Negative Stain Electron Microscopy Studies
Published: Vol 7, Iss 8, Apr 20, 2017 DOI: 10.21769/BioProtoc.2229 Views: 8197
Reviewed by: Anastasia D GaziAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Structural Characterization Using Electron Transfer Dissociation and Hydrogen Exchange-Mass Spectrometry
Rupam Bhattacharjee and Jayant B. Udgaonkar
Jun 20, 2025 1775 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 38 Views
Abstract
Legionella possesses a pivotal secretion machinery to deliver virulence factors to eukaryotic host cells. In this protocol, we describe the procedure for isolation of the native core complex of the Dot/Icm type IV secretion system from L. pneumophila aiming to perform biochemical and transmission electron microscopy analyses.
Keywords: LegionellaBackground
Legionella pneumophila is a Gram-negative bacterial pathogen that causes lung infection known as Legionnaires’ disease (Fields et al., 2002). L. pneumophila utilizes a type IV secretion system (T4SS) encoded by the dot/icm genes to transport about 300 bacterial proteins into the cytosol of their eukaryotic host to hijack cellular processes (Hubber and Roy, 2010). Composed of more than 20 proteins, the T4SS is a nanomachine built on the bacterial inner and outer membranes (Nagai and Kubori 2011; Kubori and Nagai 2016). The core complex of Dot/Icm T4SS is a biochemically stable part of the system and forms a transport conduit bridging the inner and outer membrane (Kubori et al. 2014). The core complex is composed of at least five proteins; three outer membrane-associated proteins, DotC, DotD and DotH, and two inner membrane proteins, DotF and DotG (Vincent et al., 2006). Based on the procedure for biochemical isolation of another bacterial nanomachine, the type III secretion system, from Salmonella typhimurium (Kubori et al., 1998; Marlovits et al., 2004), we modified the protocol to adapt it to the purification of the T4SS of L. pneumophla. In this protocol, we present the procedure to isolate the native core complex of the T4SS from detergent lysed wild-type L. pneumophila based on separation by ultracentrifugation. T4SS isolated using this procedure can be used to perform biochemical and transmission electron microscopy analyses described previously (Kubori et al., 2014).
Materials and Reagents
- Sterile swabs
- Sterile cell scrapers (IWAKI, catalog number: 9000-220 )
- Sterile conical tubes (50 ml and 15 ml)
- Sterile Petri dishes (100 mm in diameter)
- Cuvettes for spectrophotometer (1.5 ml) (BOECO, catalog number: BRA 759017 )
- Millex-GP filter units (EMD Millipore, catalog number: SLGP033RS )
- Sterile 10 ml syringe (Terumo, catalog number: SS-10LZ )
- Ultrafree MC filters (EMD Millipore, catalog number: UFC30GV00 )
- Sterile pipets (10 ml) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 170356 )
- L. pneumophila Lp01 strain (Philadelphia-1 rpslL hsdR) (Berger and Isberg, 1993)
- cOmpleteTM protease inhibitor cocktail (Roche Diagnostics, catalog number: 11697498001 )
- Sodium chloride (NaCl) (Nacarai Tesque, catalog number: 31320-05 )
- 12.5% precast polyacrylamide gels (ATTO, catalog number: e-PAGEL E-R12.5L ; or equivalent)
- Glow-discharged carbon grids (Nisshin EM, catalog number: 649 )
- Coomassie brilliant blue (CBB) stain One (Nacarai Tesque, catalog number: 04543-51 )
- ACES (Sigma-Aldrich, catalog number: A3594 )
- BactoTM yeast extract (BD, BactoTM, catalog number: 212750 )
- MilliQ water
- Activated charcoal (Sigma-Aldrich, catalog number: C5510 )
- BactoTM agar (BD, BactoTM, catalog number: 214010 )
- L-cysteine hydrochloride monohydrate (Nacarai Tesque, catalog number: 10313-55 )
- Iron(III) nitrate enneahydrate, Fe(NO3)3·9H2O (Nacarai Tesque, catalog number: 19514-55 )
- Tris(hydroxymethyl)aminomethane (Tris) (Nacarai Tesque, catalog number: 35434-21 )
- Hydrochloric acid (HCl) (Nacarai Tesque, catalog number: 18321-05 )
- Sucrose (Nacarai Tesque, catalog number: 30404-45 )
- Phenylmethylsulfonyl fluoride (PMSF) (Nacarai Tesque, catalog number: 27327-94 )
- Isopropanol (Sigma-Aldrich, catalog number: 190764 )
- EDTA·2Na (Nacarai Tesque, catalog number: 15130-95 )
- Sodium hydroxide (NaOH) (Nacarai Tesque, catalog number: 31511-05 )
- Lysozyme (Wako Pure Chemical Industries, catalog number: 120-02674 )
- Triton X-100 (Nacarai Tesque, catalog number: 35501-15 )
- AG501-X8 Resin (Bio-Rad Laboratories, catalog number: 143-7425 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Nacarai Tesque, catalog number: 21003-75 )
- DNase I (Sigma-Aldrich, catalog number: DN25 )
- Potassium hydroxide (KOH) (Nacarai Tesque, catalog number: 28616-45 )
- Uranyl acetate (UA) (Merck, catalog number: 8473 )
- Phosphotungstic acid (PTA) (TAAB, catalog number: p013 )
- CYE plate (see Recipes)
- AYE medium (see Recipes)
- Tris-Cl solution (pH 8.0) (see Recipes)
- Sucrose solution (see Recipes)
- PMSF stock solution (see Recipes)
- EDTA stock solution (see Recipes)
- Lysozyme solution (see Recipes)
- Triton X-100 stock solution (see Recipes)
- MgSO4 stock solution (see Recipes)
- DNase I stock solution (see Recipes)
- KOH solution (see Recipes)
- NaOH stock solution (see Recipes)
- TET solution (see Recipes)
- PTA solution (see Recipes)
- Uranyl acetate solution (see Recipes)
Equipment
- Glass flasks (2 L)
- 37 °C shaking incubator
- 37 °C incubator
- Spectrophotometer
- Refrigerated centrifuge (KUBOTA, model: 7780 ; or equivalent models)
- Rotor for centrifugation (KUBOTA, models: AG-5006 , AG-6512C )
- Sterile centrifuge tubes (500 ml capacity, polypropylene or polycarbonate)
- Sterile centrifuge tubes (50 ml capacity, polyallomer or polycarbonate)
- Clean grass beakers (200 ml)
- Magnetic stir bars
- Magnetic stirrer
- pH meter
- Ultracentrifugation (Beckman Coulter, model: OptimaTM L-100 XP ; or equivalent models)
- Rotor for ultracentrifugation (Beckman Coulter, model: Type 70Ti )
- Tubes for ultracentrifugation (Beckman Coulter, catalog number: 355631 )
- Microfuge (Eppendorf, model: 5415 R )
- Glass beakers (1 L)
- Autoclavable flasks (2 L)
- Autoclave
- Electrophoresis apparatus (ATTO, model: AE-6530 ; or equivalent model)
- Electron microscope (JEOL, model: JEM-1011 )
- ÄKTA purifier (GE Healthcare)
- Superose 6 10/300 GL column (GE Healthcare, catalog number: 17517201 )
Procedure
- Grow L. pneumophila Lp01 strain on a charcoal-yeast extract (CYE) plate (Recipe No. 1) from a glycerol frozen stock for 3 days at 37 °C.
- Use a sterile swab to streak a L. pneumophila colony on the entire surface of CYE plates and cultivate for 2 days at 37 °C.
Note: Two plates covered with a bacterial lawn are needed to obtain sufficient starting material to prepare 1 L culture. - Take the whole bacteria using sterile cell scrapers and suspend it with ~5 ml ACES-buffered yeast extract (AYE) medium (Recipe No. 2) in a sterile tube. Measure OD600. Add the bacterial solution into 1 L AYE medium in a 2 L flask to make the suspension of OD600 0.2.
- Grow the bacteria for 12 h at 37 °C with rotary shaking (250 rpm).
- Chill the flask in icy water. Measure OD600. (It should be 2.0-3.0, indicating the late log phase of growth). Transfer the culture solution to chilled centrifuge tubes (2-3 tubes of 500 ml capacity).
- Harvest the bacteria by centrifugation at 12,000 x g (8,000 rpm) for 15 min at 4 °C (Refrigerated centrifuge: Kubota, model: 7780 or equivalent models; Rotor: Kubota, model: AG-5006).
- Resuspend the bacterial pellet with 140 ml of cold sucrose solution (Recipe No. 4) containing 1x cOmpleteTM protease inhibitor cocktail.
Note: Thoroughly suspending bacterial pellets enhances the efficiency of the detergent lysis in step 11. - Transfer the suspension to a 200 ml beaker with a magnetic stir bar on ice.
- Set the beaker on a magnetic stirrer at room temperature, and stir mildly until uniform suspension is achieved.
- Add PMSF (final 1 mM, Recipe No. 5), EDTA (final 1 mM, Recipe No. 6) and lysozyme (final 0.1 mg/ml, Recipe No. 7) in this order.
Note: This is the process of spheroplast formation.
Prepare a 20 mg/ml 200x stock solution. Add 0.7 ml of this stock solution to the 140 ml sucrose solution, yielding a final lysozyme concentration of approximately 0.1 mg/ml in the bacterial resuspension. Keep stirring for another 30 min at room temperature. - Add slowly (drop by drop) 7 ml of 20% (w/v) Triton X-100 stock solution (final 1% w/v, Recipe No. 8) with stirring.
Note: This is the most important step for detergent lysis of bacterial membranes. Carefully monitor the change of color and viscosity. - Keep stirring until the solution becomes very clear (approximately for 30 min).
- Add 420 μl of 1 M MgSO4 stock solution (final 3 mM, Recipe No. 9) and 70 μl of 10 mg/ml DNase I stock solution (final 5 μg/ml, Recipe No. 10) in this order.
Note: This step is required to digest DNA. - Stir for 10 min.
- Add EDTA (final 10 mM).
- Monitoring pH using a pH meter, adjust pH to 10.0 by adding 1 N NaOH (Recipe No. 12).
Note: During this step, large membrane vesicles and fragments that remain are disrupted. The core complex is stable at this high pH, while loosely associated contaminant proteins can be detached from the core complex. For the purpose of analyzing the loosely associated components of the T4SS, pH value can be modified to maintain these proteins in the complex. - Move the beaker on ice.
Note: Procedure below should be done at 4 °C.
- Transfer the lysate to chilled 50 ml centrifuge tubes (divide into 3 tubes).
- Centrifuge the lysate at 12,000 x g for 20 min at 4 °C to remove non-lysed materials. (Refrigerated centrifuge: Kubota, model: 7780 or equivalent models; Rotor: Kubota, model: AG-6512C).
- Recover supernatant and transfer to ultracentrifuge tubes (Beckman Coulter).
- Apply ultracentrifugation at 100,000 x g for 30 min at 4 °C to precipitate protein complexes. (Ultracentrifuge: Beckman Coulter, model: Optima L-100 XP or equivalent models; Rotor: Beckman Coulter, model: Type 70Ti)
- Discard supernatant. Soak the pellet with 0.5 ml of cold TET solution (Recipe No. 13) including 1 mM PMSF per tube. To dissolve completely, leave the pellet in TET overnight at 4 °C.
- Merge the completely dissolved suspensions and transfer them in a new conical or centrifuge tube (total ~5 ml). Use extra 2-3 ml of cold TET solution to completely recover the dissolved proteins from the tubes and merge them into the new tubes.
- Centrifuge the suspension at 14,000 x g for 15 min at 4 °C to remove precipitate. (Refrigerated centrifuge: Kubota, model: 7780 or equivalent models; Rotor: Kubota, model: AG-6512C).
- Submit the supernatant to a second round of ultracentrifugation at 100,000 x g for 30 min at 4 °C. (Ultracentrifuge: Beckman Coulter, model: Optima L-100 XP or equivalent models; Rotor: Beckman Coulter, model: Type 70Ti)
- Resuspend the pellet in ~300 μl of cold TET. The sample is ready for biochemical and electron microscopic analyses (Figures 1A and 1B).
- (Optional) Further separation by Superose 6 10/300 column chromatography equilibrated with TET plus 50 mM NaCl can be used to remove large aggregates either made of T4SS or contaminant proteins. Collect the fractions and identify the fractions containing the core complex by gel electrophoresis and transmission electron microscopy (as shown in steps 28 and 29) (Figures 1C and 1D).
Note: As TET solution contains Triton X-100 that disturbs UV monitoring, the UV peaks do not always accord with the presence of the protein complexes. Alternative approaches like sucrose density gradient ultracentrifugation can be applicable for further purification. - For biochemical analysis, apply samples on 12.5% SDS-polyacrylamide gel (PAGE) and stain with ready-made Coomassie Brilliant Blue stain solution, CBB stain One (Nacarai).
- For transmission electron microscopy analysis, apply samples on glow-discharged carbon grids and negatively stained with 2% (w/v) PTA pH 7.0 (Recipe No. 14) or 2% (w/v) uranyl acetate (Recipe No. 15). Take micrographs at an accelerating voltage of 80 kV.
Data analysis
The second round of ultracentrifugation enhances the purity of the isolated complex, and the following column chromatography further removes the background contaminations (Figure 1).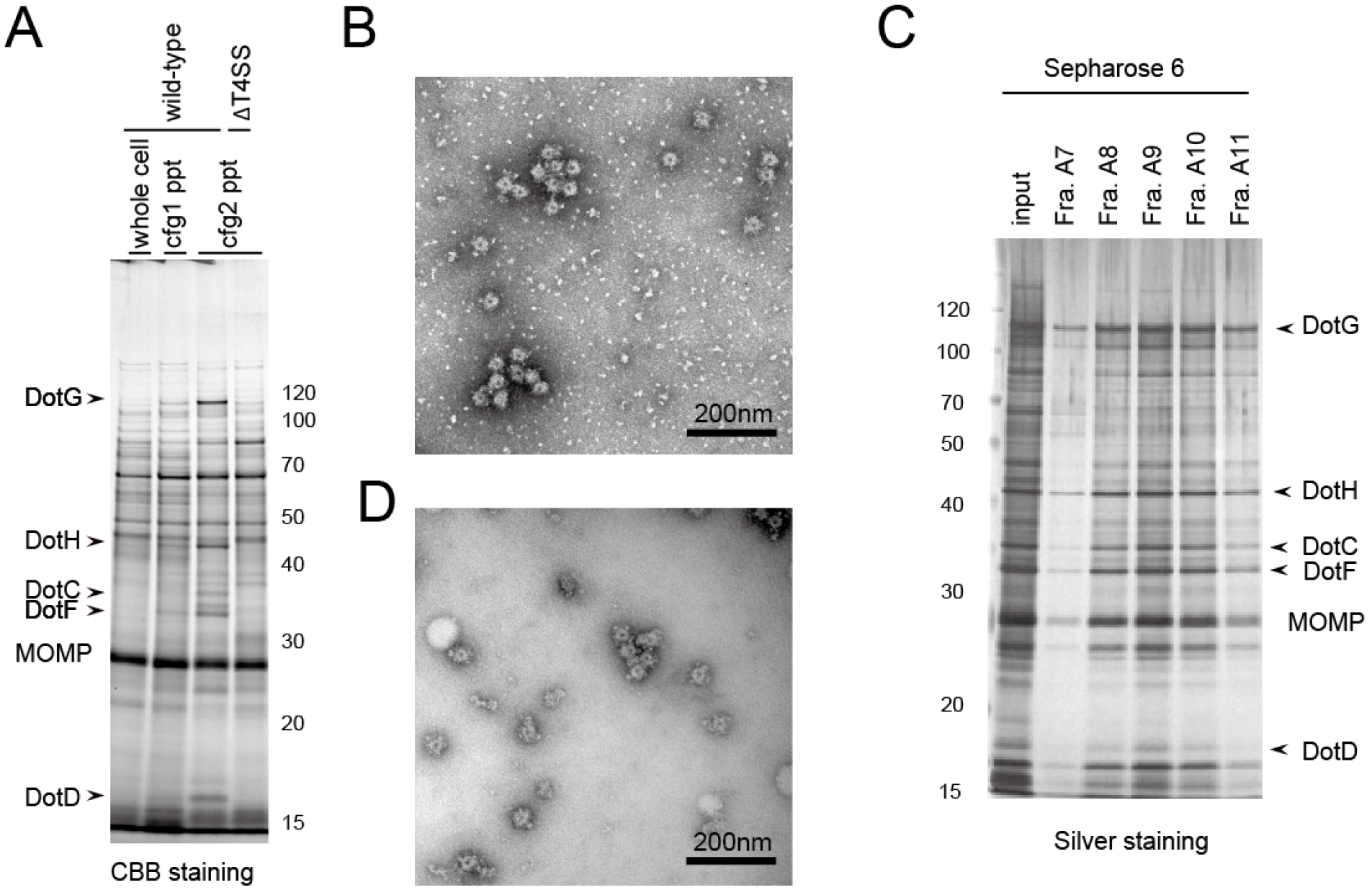
Figure 1. Example of isolated Dot/Icm T4SS core complex. A. SDS-PAGE analysis of the isolated complex in comparison between 1st and 2nd rounds of ultracentrifugation (steps 21 and 25, respectively). Whole cell: Whole cell lysate (step 20); cfg1 ppt: 1st ultracentrifugation pellet (step 22); cfg2 ppt: 2nd ultracentrifugation pellet (step 26). As a negative control, a L. pneumophila strain lacking all dot/icm genes (∆T4SS) was also submitted to the isolation procedure (the rightmost lane). B. The electron micrograph of the fraction obtained by the 2nd ultracentrifugation; C. SDS-PAGE analysis of the fractions obtained by a size exclusion column chromatography (step 27); D. The electron micrograph of the fraction A10 of (C). Molecular weight markers are shown in kDa. MOMP: Major Outer membrane Protein. The images are adapted from Kubori et al. (2014).
Recipes
- CYE plate (for 24 plates, 1 L)
10 g ACES
10 g yeast extract
Dissolve in ~0.9 L MilliQ water in a 1 L glass beaker, adjust pH by adding 1 N KOH (see Recipe No. 11; ~40 ml) and monitoring with pH meter
Bring the volume up to 1 L
Add to a 2 L autoclavable flask containing a magnetic stirrer bar and 2 g activated charcoal and 15 g agar
Mix briefly by stirring
Autoclave the media at 121 °C for 30 min
Cool down at room temperature until the temperature reaches ~60 °C with gentle stirring. During the time, prepare L-cysteine (0.4 g solved in 10 ml sterile MilliQ water) and Fe(NO3)3 (0.135 g solved in 10 ml sterile MilliQ water) solutions in sterile 15 ml conical tubes, and filter them with Millex-GP filter unit with 10 ml syringe
Add L-cysteine and Fe(NO3)3 solutions (10 ml each) to 1 L medium with stirring
Keep stirring for another 5 min to mix homogeneously
Pour ~40 ml per Petri dish and cool down to solidify - AYE medium (1 L)
Same as CYE except that agar is omitted
Note: AYE is not good for use on the same day of preparation. For good result, AYE should be made one day before the Legionella culture. Old media (more than a week after preparation) is not recommended. - Tris-Cl solution (pH 8.0)
1 M solution
Adjust pH to 8.0 with HCl
Autoclave at 121 °C for 20 min - Sucrose solution
0.5 M sucrose
150 mM Tris-Cl (pH 8.0) - PMSF stock solution
100 mM in isopropanol
Storage at -20 °C - EDTA stock solution
0.5 M EDTA
Adjust pH to 8.0 with NaOH
Autoclave at 121 °C for 20 min - Lysozyme solution
20 mg/ml in sucrose solution
Prepare just before use - Triton X-100 stock solution
20% (w/v) solution containing ~2 g of AG501-X8 Resin for deionizing - MgSO4 stock solution
1 M solution
Autoclave at 121 °C for 20 min - DNase I stock solution
Dissolve the powder in sterile water to give 10 mg/ml solution
Storage at -20 °C - KOH solution
1 N solution - NaOH stock solution
1 N solution - TET solution
10 mM Tris-Cl (pH 8.0)
1 mM EDTA
0.1% Triton X-100 - PTA solution
2% (w/v) phosphotungstic acid. Adjust to pH 7.0 - Uranyl acetate solution
2% (w/v) uranyl acetate
Filter with 0.22 μm ultra-free MC
Acknowledgments
This work has been financially supported by MEXT/JSPS KAKENHI Grants 15H01322 (to TK). The protocol presented here has been adapted from Kubori et al. (2014).
References
- Berger, K. H. and Isberg, R. R. (1993). Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7(1): 7-19.
- Fields, B. S., Benson, R. F. and Besser, R. E. (2002). Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev 15(3): 506-526.
- Hubber, A. and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261-283.
- Kubori, T., Koike, M., Bui, X. T., Higaki, S., Aizawa, S. and Nagai, H. (2014). Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc Natl Acad Sci U S A 111(32): 11804-11809.
- Kubori, T., Matsushima, Y., Nakamura, D., Uralil, J., Lara-Tejero, M., Sukhan, A., Galan, J. E. and Aizawa, S. I. (1998). Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280(5363): 602-605.
- Kubori, T. and Nagai, H. (2016). The Type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol 29: 22-29.
- Marlovits, T. C., Kubori, T., Sukhan, A., Thomas, D. R., Galan, J. E. and Unger, V. M. (2004). Structural insights into the assembly of the type III secretion needle complex. Science 306(5698): 1040-1042.
- Nagai, H. and Kubori, T. (2011). Type IVB secretion systems of legionella and other Gram-negative bacteria. Front Microbiol 2: 136.
- Vincent, C. D., Friedman, J. R., Jeong, K. C., Buford, E. C., Miller, J. L. and Vogel, J. P. (2006). Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 62(5): 1278-1291.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kubori, T. and Nagai, H. (2017). Isolation of the Dot/Icm Type IV Secretion System Core Complex from Legionella pneumophila for Negative Stain Electron Microscopy Studies. Bio-protocol 7(8): e2229. DOI: 10.21769/BioProtoc.2229.
- Kubori, T., Koike, M., Bui, X. T., Higaki, S., Aizawa, S. and Nagai, H. (2014). Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc Natl Acad Sci U S A 111(32): 11804-11809.
Category
Microbiology > Microbe-host interactions > Bacterium
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










