- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Assay to Assess Efficacy of Potential Antiviral Compounds against Enterovirus D68
Published: Vol 7, Iss 6, Mar 20, 2017 DOI: 10.21769/BioProtoc.2183 Views: 10397
Reviewed by: Yannick DebingKristin ShinglerCristina SuárezNoam Vardi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Production and Purification of Cell Culture–generated Hepatitis B Virus by Transient Transfection and Density Gradient
Asako Murayama [...] Takanobu Kato
Jul 20, 2023 1934 Views

An Improved Focus-Forming Assay for Determination of the Dengue Virus Titer
Maharah Binte Abdul Mahid [...] Kitti Wing Ki Chan
Oct 20, 2024 2444 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2475 Views
Abstract
In 2014 enterovirus D68 (EV-D68) caused the largest outbreak in the United States since the discovery of the virus. Distinct from before, the 2014 infections were associated with more severe respiratory disease and occasional neurological complications. So far, there are no available vaccines or antivirals for the prophylaxis or treatment of EV-D68 infections. In order to evaluate the antiviral activity of potential inhibitors of EV-D68 replication, a cell-based cytopathic effect (CPE) reduction assay was developed (Sun et al., 2015).
Keywords: Enterovirus D68Background
As a re-emerging pathogen, antiviral compounds targeting EV-D68 were rarely reported before. It is urgently needed to establish and develop antiviral methods to combat the potential EV-D68 epidemic. Here, we report a detailed protocol which can be used to identify selective anti-EV-D68 compounds. To screen and identify potential antiviral compounds, the MTS-based CPE reduction assay is reproducible, easy-to-use and time-saving method which is widely used. This method relies on the ability of a compound to inhibit EV-D68-induced reduction of CPE. When a compound actively inhibits the replication of EV-D68, the virus-induced CPE will be reduced or absent. As a consequence, the metabolically active cells are able to convert a yellow tetrazolium salt substrate (MTS/PMS) to a brown-colored formazan product. When a compound does not have antiviral effects, the host cells die of virus-induced CPE, which will lack metabolic activity, and the yellow substrate remains un-metabolized. The colorimetric conversion is quantitative, as an EC50 can be calculated from the data generated by this assay.
Materials and Reagents
- Centrifuge tube, 14 ml (Corning, Falcon®, catalog number: 352059 )
- 1.5 ml Eppendorf Snap-Cap microcentrifuge tubes (VWR, catalog number: 21008-959 )
- 4 ml Screw neck vials, amber glass (VWR, catalog number: 548-0052 )
- Transparent 96-well (flat-bottom) tissue culture plates (Corning, Falcon®, catalog number: 353072 )
- Tissue culture flasks, 150 cm2 (TPP, catalog number: 90856 )
- Pipette tips (10 µl, 100 µl, 1,000 µl)
- Disposable serological pipets (5 ml, 10 ml and 25 ml)
- Reagent reservoir, 25 ml (Biotix, catalog number: SR-0025-5SWM )
- Nalgene rapid-flow sterile disposable filter units with SFCA membrane (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 156-4020 )
- Hela Rh cell line (a Hela subclone, highly susceptible and permissible to rhinovirus-induced CPE, a gift from Janssen Pharmaceutica, Belgium) (Lacroix et al., 2014)
- All EV-68 strains were cultivated on HeLa Rh cells and stored at -80 °C
- EV-D68 strains (742, 947, 2042, 1348, 2284 and 670) were isolated from the Netherlands (a kind of gift from Adam Meijer, Diagnostics and Screening National Institute for Public Health and the Environment [RIVM], Bilthoven)
- EV-D68 strains (US/KY/14-18953, US/IL/14-18952 and US/MO14-18947) – these strains were originally obtained from BEI Resources
- EV-D68 strains (742, 947, 2042, 1348, 2284 and 670) were isolated from the Netherlands (a kind of gift from Adam Meijer, Diagnostics and Screening National Institute for Public Health and the Environment [RIVM], Bilthoven)
- Dulbecco’s phosphate-buffered saline (DPBS), no calcium, no magnesium (Thermo Fisher Scientific, GibcoTM, catalog number: 14190144 )
- 0.05% trypsin-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25300054 )
- Minimum essential media (MEM) Rega-3 (Thermo Fisher Scientific, GibcoTM, catalog number: 19993013 )
- Potential antiviral compounds
- SG85*
- Pleconaril*
- Vapendavir*
- Pirodavir*
- Rupintrivir*
- Enviroxime*
- Favipiravir*
- SG85*
- Fetal bovine serum (GE Healthcare, HycloneTM, catalog number: SH30084.03 )
- L-glutamine 200 mM (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Sodium bicarbonate 7.5% solution (Thermo Fisher Scientific, GibcoTM, catalog number: 25080060 )
- Minimum essential media (MEM), no glutamine, no phenol red (Thermo Fisher Scientific, GibcoTM, catalog number: 51200038 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- CellTiter 96® AQueous MTS reagent powder (Promega, catalog number: G1111 )
- HCl
- Dimethyl sulfoxide (VWR, catalog number: 67-68-5 )
- Growth culture medium (see Recipes)
- Assay medium (see Recipes)
- MTS/PMS solution (see Recipes)
Equipment
- Pipette controller, PIPETBOY acu2 (VWR, catalog number: 612-0927 )
- Multichannel pipette, 10-100 µl (Eppendorf)
- Incubator at 37 °C (and 35 °C ) and 5% CO2 (Binder)
- Mid bench centrifuge (Sigma Laborzentrifugen, mode: 4K15C )
- MoxiTM Z Mini Automated Cell Counter (ORFLO Technologies, catalog number: MXZ001 )
- SpectraMax 190 microplate reader (Molecular Devices, model: SpectraMax 190 )
- pH bench meter (Xylem Analytics, WTW, model: inoLab® 9310 IDS )
- Inverted microscope (Motic, model: AE21 )
- Safety cabinet (Telstar)
Software
- GraphPad Prism
Procedure
- Grow Hela Rh cells to confluence in growth culture medium (see Recipes) at 37 °C and 5% CO2.
Note: Hela Rh cells are passaged by 1 in 10, and they will be confluence after 3-4 days. - Aspirate the supernatant, wash the cells three times with 10 ml PBS and detach the cells using trypsin at 37 °C about 3-5 min.
- When the cells are detached, collect the cells with 10 ml growth medium in a 14 ml tube. Centrifuge the cells for 6 min at 269 x g at room temperature.
- Re-suspend the cell pellet in 10 ml assay medium (see Recipes).
- Count the number of cells. Seed the cells into a 96-well plate at a density of 15,000 cells in 100 µl assay medium per well. Incubate the cells for 24 h in the incubator at 37 °C and 5% CO2 to allow the cells to adhere to the plate.
- Dilute the compounds to the desired start concentration in assay medium. This start concentration should be 6x the desired final concentration to prepare for the 1-in-3 serial dilution. Add 50 µl of the 6x compound dilution to the 100 µl assay medium in the second column, wells B2 to G2 which seeded with cells (a maximum of 6 compounds, Figure 1).
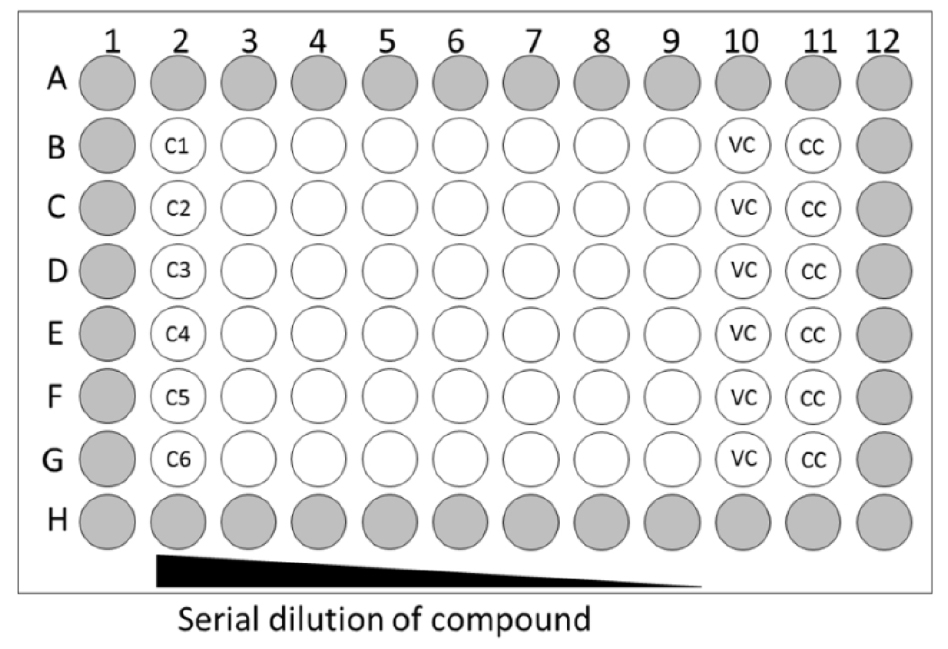
Figure 1. Layout of 96-well plate for antiviral assay. Grey wells only contain medium; VC: virus control, CC: cell control. - Mix the medium of column 2 by gentle pipetting and transfer 50 µl of medium from this column to the next column 3 (B3-G3) (see Figure 2A); repeat this up to column 9 (B9-G9). Discard 50 µl of medium from column 9. Column 10 (B10-G10) will be the virus control (VC, assay medium + EV68) and column 11 (B11-G11) will be the cell control (CC, only assay medium).
- Add 100 µl of EV68 of the desired dilution (MOI = 0.001) to column 2-10 and 100 µl assay medium to column 11 (see Figure 2B). Wells on the edge of the 96-well plate (see Figure 1, labelled in grey) contain 200 µl medium but will not be used because edge effects may influence the assay results.
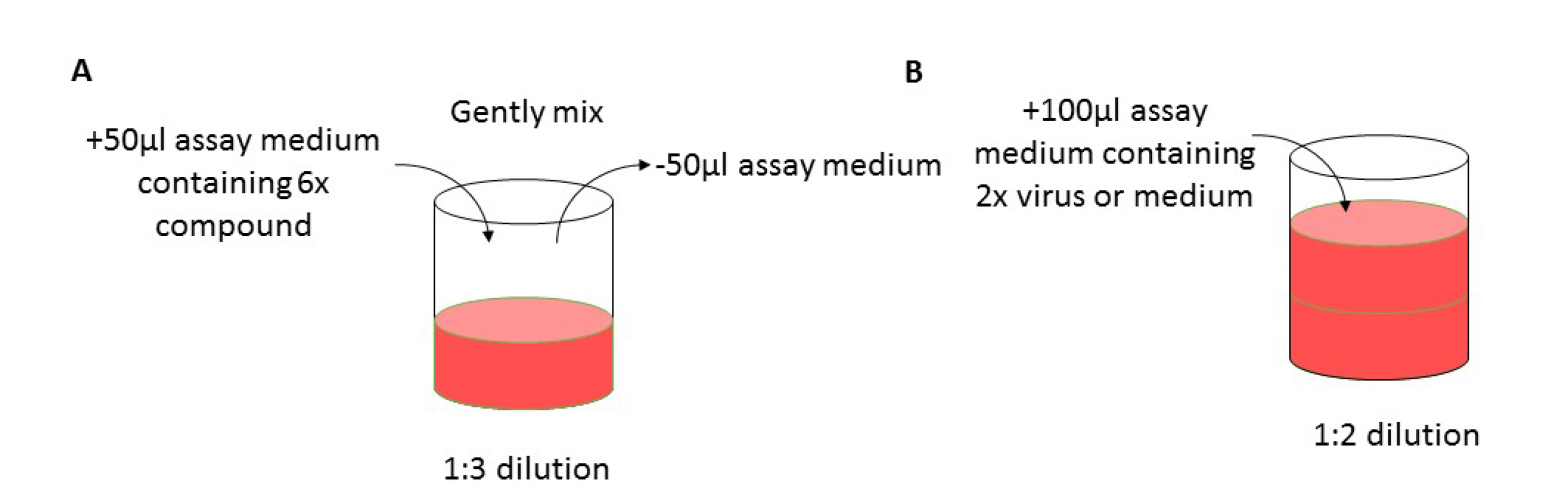
Figure 2. Workflow. A. Gently mix the assay medium in the treated and infected wells. Transfer 50 µl of medium to the next well to prepare a 1-in-3 dilution series. B. Following the preparation of compound dilutions, add 100 µl of assay medium containing 2x virus dilution. - Place the 96-well plate in the incubator (35 °C, 5% CO2 and 95% relative humidity) for 3-4 days until complete virus-induced cytopathic effect (CPE) is observed by microscope in the VC wells (see Figure 3).
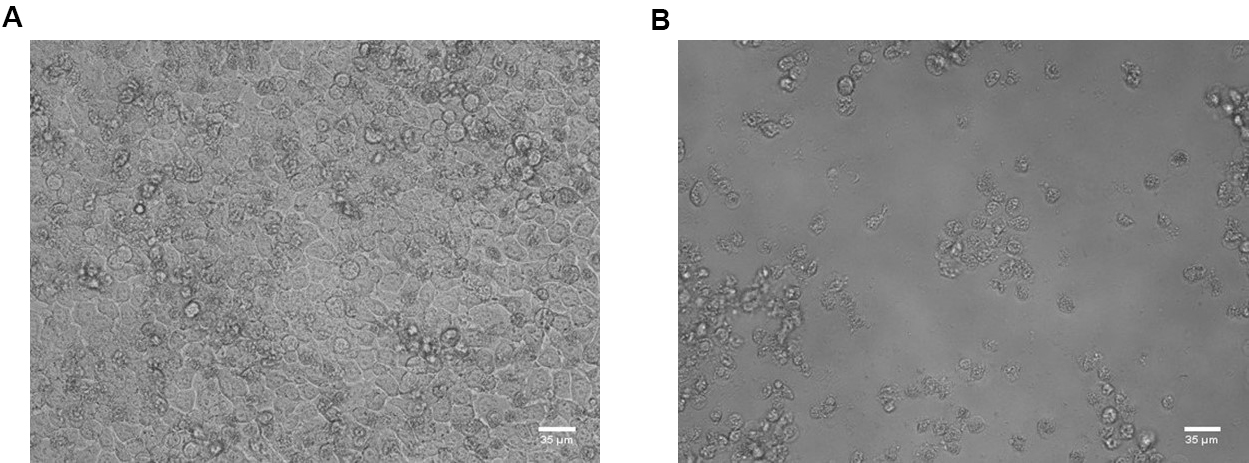
Figure 3. HeLa Rh cells in cell control wells (A) and virus control wells (B). Scale bars = 35 μm. - Remove the assay medium of all wells in column 2-11 and add 100 µl of 5% MTS/PMS dilution in phenol red- and glutamine-free medium.
- Following incubation for 40-60 min in the 37 °C 5% CO2 incubator, measure the absorbance in each well of the 96-well plate with a microplate reader at a wavelength of 498 nm.
Data analysis
- The antiviral activity is calculated as follows:
% antiviral activity = (ODEV68+Compound - ODVC)/(ODCC - ODVC) x 100.
Where,
ODCC and ODVC represent average values of optical density (OD) of the cell control (column 11) and the virus control (column 10), respectively,
ODEV68+Compound corresponds to the OD value of each infected and treated well. - Calculate the EC50 value of each compound, which is defined as the concentration of compound at which the virus-induced CPE is reduced by 50%. This value of antiviral activity is calculated by curve fitting which was generated from GraphPad Prism.
Note: Alternatively, logarithmic interpolation can be used to estimate EC50 values if GraphPad Prism is not available. The below Representative data is shown by logarithmic interpolation.
Representative data
- % Antiviral activity of each concentration is calculated and showed below:
SG85 (µM) 0.0002 0.0005 0.002 0.005 0.014 0.043 0.13 0.38 % antiviral activity 2.35 5.32 13.85 91.50 98.86 93.33 101.78 104.03
EC50 =10((Log(A) – Log(B)) x ((C - 50)/(C - D)) + Log(B))
Where,
A: compound concentration at which % antiviral activity is < 50%
B: compound concentration at which % antiviral activity is > 50%
C: value of % antiviral activity > 50%
D: value of % antiviral activity < 50% - An example of a dose-response curve of the antiviral activity of the protease inhibitor SG85 against different EV68 clinical isolates (see Figure 4). EC50 values are presented in Sun et al. (2015).
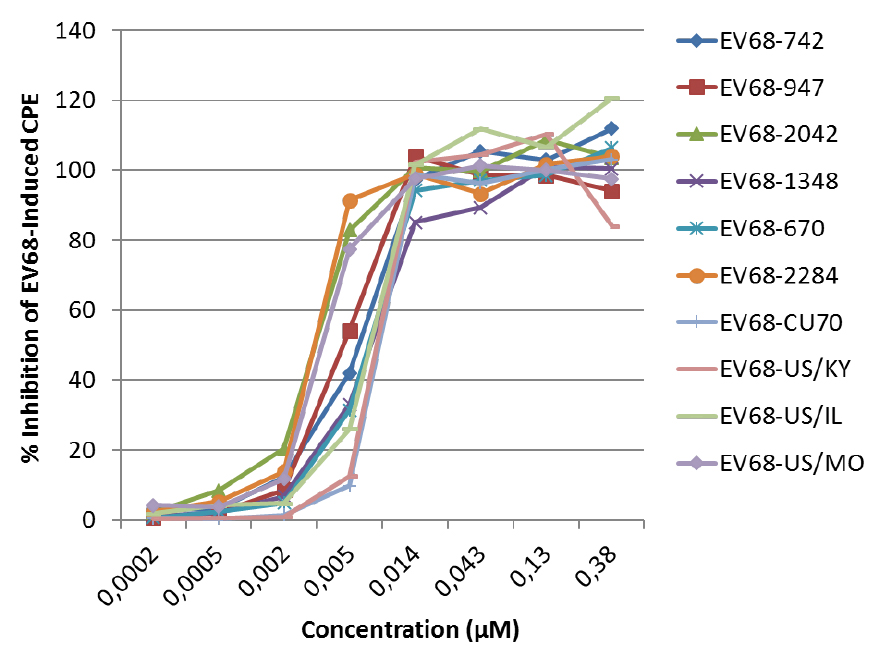
Figure 4. The antiviral activity of SG85 against EV68 clinical isolates was determined in a CPE reduction assay with an MTS/PMS readout
Notes
- Depending on the cell type used in the antiviral assay, the amount of cells per well may need to be optimized. It is important that cells in the cell control group do not over-grow after 3 or 4 days.
- Use the lowest viral inoculation that still results in 100% full CPE in all wells at day 3 or 4 post infection.
- If a 1-to-2 serial dilution of compound is preferred, prepare a 4x concentrated compound dilution and add 100 µl of this compound dilution to column 2; Then transfer 100 µl from well to well to generate the 1:2 dilution series.
- Because of its low optimum growth temperature, EV-D68 is propagated well at 35 °C.
- The time of incubation of the MTS dilution depends on the cell type and cell density. The cells should stay in the incubator until a brown color appears in the CC wells (B11-G11) after the MTS solution has been added. The OD value of the CC should be between 0.6 and 1 to stay in the linear range. The incubation time is approximately 40 to 60 min for Hela Rh cells at 37 °C.
- The absorbance value of brown-colored formazan product can reach the biggest at wavelength of 498 nm.
- In general, this assay/method is adaptable for other picornaviruses with a few considerations (e.g., cell types, medium and temperature).
Recipes
- Growth culture medium
MEM supplemented with:
10% fetal calf serum
2 mM L-glutamine
0.075% sodium bicarbonate - Assay medium
MEM supplemented with:
2% fetal calf serum
2 mM L-glutamine
0.075% sodium bicarbonate
30 mM MgCl2 - MTS/PMS solution
- 1 g of MTS powder is dissolved in 500 ml of PBS and stirred
- Adjust pH of solution with 1 N HCl to be between 6.0-6.5
- 23 mg of PMS powder is added until complete dissolution by stirring
- Filter the solution through a rapid-flow sterile disposable filter unit
- 3 ml aliquot solution is stored at -20 °C (it should be kept in the dark to avoid degradation)
Acknowledgments
This work was supported by a fellowship to Liang Sun from China Scholarship Council (CSC) (grant 201403250056) and the Interuniversitaire attractiepolen (IUAP) BELVIR. The protocol described here was based on the following paper: Sun et al. (2015).
References
- Lacroix, C., Querol-Audi, J., Roche, M., Franco, D., Froeyen, M., Guerra, P., Terme, T., Vanelle, P., Verdaguer, N., Neyts, J. and Leyssen, P. (2014). A novel benzonitrile analogue inhibits rhinovirus replication. J Antimicrob Chemother 69(10): 2723-2732.
- Sun, L., Meijer, A., Froeyen, M., Zhang, L., Thibaut, H. J., Baggen, J., George, S., Vernachio, J., van Kuppeveld, F. J., Leyssen, P., Hilgenfeld, R., Neyts, J. and Delang, L. (2015). Antiviral activity of broad-spectrum and enterovirus-specific inhibitors against clinical isolates of enterovirus D68. Antimicrob Agents Chemother 59(12): 7782-7785.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sun, L., Delang, L., Mirabelli, C. and Neyts, J. (2017). In vitro Assay to Assess Efficacy of Potential Antiviral Compounds against Enterovirus D68. Bio-protocol 7(6): e2183. DOI: 10.21769/BioProtoc.2183.
Category
Microbiology > Antimicrobial assay > Antiviral assay
Cell Biology > Cell-based analysis > Viral infection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










