- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Resin-embedded Thin-section Immunohistochemistry Coupled with Triple Cellular Counterstaining
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2052 Views: 8211
Reviewed by: Tie LiuGaston A. PizzioFang Xu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2312 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1688 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 749 Views
Abstract
This protocol was developed to study protein localisation within the vascular bundles of developing tomato fruit however, it can be applied to any resin embedded plant tissue. The vascular bundle is comprised of many different cells that all have unique properties. The mature sieve elements are enucleated and contain sieve plates that comprise of callose. This method has utilised these properties of the sieve element by combining immunohistochemistry for cell wall invertase with counterstaining of aniline blue for callose, DAPI for nucleus and cell structure is shown with the final staining of the cell wall using calcofluor white. It must be noted that when following this protocol, it is vital for the sections to be flat and fixed to the slide with gelatine so cover slip removal does not move the sample section. This protocol will be applicable to all plant tissues and provides additional evidence of the protein localisation within the cell by conducting a counterstaining procedure.
Keywords: ImmunolocalisationBackground
Immunolocalisation has long been a method used to study the localisation of proteins within tissue. This protocol focused on, not only localising the proteins of interest but also the molecular structures that were in surrounding tissue. It is believed that the mature sieve elements are enucleated and have abundant callose deposition within the sieve plate. Therefore, counterstaining procedures were applied in order to represent these biological phenomena. As the proteins of interest in this study were also thought to be localised within the apoplast further counterstaining was applied showing co-labelling of the protein and the cell wall.
Materials and Reagents
- Size 000 gelatine capsules (ProSciTech, catalog number: RL039 )
- Razor blade
- Microscope slide (Livingstone, catalog number: 7107-PPN )
- 22 x 50 mm, 0.17 mm thick coverslip (Fisher Scientific, catalog number: 12-543C )
- Tomato flowers 2 Days Before Fertilization (DBF) and 2 Days After Fertilization (DAF) harvested from glasshouse-grown cv. Moneymaker tomato plants
- 50 mM PIPES (Sigma-Aldrich, catalog number: F6757 )
- AR grade EtOH (Sigma-Aldrich, catalog number: 32205 )
Note: This product has been discontinued. - ddH2O
- LR white resin (ProSciTech, catalog number: C023 )
- Gelatine (Sigma-Aldrich, catalog number: G9391 )
- Cell wall invertase (LIN5) purified polyclonal primary antibody – produced in rabbit (Mimmitopes – custom made)
- Inhibitor of invertase (INH) purified polyclonal primary antibody – produced in rabbit (Mimmitopes – custom made)
- TBST (Sigma-Aldrich, catalog number: T9039 )
- Secondary anti-rabbit IgG fluorescein isothiocyanate (FITC) (Sigma-Aldrich, catalog number: F9887 )
- Anti-Rabbit IgG (whole molecule)-FITC antibody produced in goat (Sigma-Aldrich, catalog number: F6005 )
Note: This product has been discontinued. - Aniline blue (0.1% in ddH2O) (Sigma-Aldrich, catalog number: B8563 )
- Mowiol-phenylenediamine (mowiol) (Sigma-Aldrich, catalog number: 10852 )
- 4’,6-diamidino-2-phenylindole (DAPI) (1:500) (Sigma-Aldrich, catalog number: D9542 )
- Calcofluor white (0.1% in ddH2O) (Sigma-Aldrich, catalog number: F3543 )
- 2% paraformaldehyde
- Glutaraldehyde
- CaCl2
- Tris
- NaN3
- Bovine serum albumin (BSA)
- Fixing solution (see Recipes)
- Blocking buffer (see Recipes)
Equipment
- Rotator
- Dissecting microscope or magnifying glass
- Reichert Ultracut E microtome (Reichert, model: 701704 )
- DiATOME Histo Knife, Diamond, 45°, 4.0-4.9 mm (ProSciTech, catalog number: UH45-40 )
- Fume hood
- Beaker
- Axio Scope.A1 epifluorescence compound microscope (ZEISS, model: Axio Scope.A1 )
- Emission FITC filter (50-490 nm excitation, long pass 515 nm)
- Emission UV filter (365 nm excitation, short pass 420 nm)
- AxioCam digital camera (ZEISSTM) or equivalent
Software
- AxioVision V4.8 software
- Adobe Bridge CS4 software
- Abode Photoshop CS4 software
Procedure
- Sample preparation
- Harvest whole tomato flowers 2 DBF and 2 DAF from glasshouse-grown tomato plants and placed on ice for 4 h in fixing solution under vacuum (300 mm Hg) to remove air bubbles and submerge the entire flower.
- Wash flowers 3 x 10 min in 50 mM PIPES and store overnight at 4 °C.
- Was a further 3 x 10 min in ddH2O preceding ethanol dehydration.
- Ethanol dehydration series is progressed 10-100% in 10% increments whilst flowers are submerged in 5 ml of solution for 1 h per increment at 4 °C up to 70% then store tissue overnight.
- The following day remaining increments 80% and 90% are applied for 1 h at RT. Once in 100% ethanol, flowers are washed twice in 100% at 1 h increments before again being stored overnight.
- LR white resin is diluted with ethanol and infiltration series conducted as above with LR white concentrations starting at 10% through to 100% on a rotator at RT.
- Once samples are in 100% LR white resin, change with 5 ml fresh 100% LR white solution every 2-3 days for a period of three weeks.
- Size 000 gelatine capsules are 1/3 filled with LR white resin and partly polymerised at 60 °C (~12 h). Flowers are then placed into middle of each capsule with petals pointing up, on the partly polymerised resin and filled with LR white resin.
- The gelatine capsules are sealed and placed in 60 °C oven for 48 h until resin is totally polymerised.
- Hardened capsules are then trimmed with a razor blade around each flower sample under a dissecting microscope leaving 1 mm resin around all surfaces.
- The trimmed blocks are securely fastened onto a Reichert Ultracut E microtome and 1 µm thick sections cut using 45 Diamond DiATOME Histo Knife, 4.0-4.9 mm.
- Sections are floated onto water at RT within the knife boat before being transferred to a drop of ddH2O on a gelatine coated LIVINGSTONETM microscope slide.
- Three sections are placed on each slide and dried on a laboratory warming tray at 45 °C in fume hood under a beaker with a chloroform soaked cotton wool bud attached to the underside until water drop evaporated and the section flattens onto the slide.
- Harvest whole tomato flowers 2 DBF and 2 DAF from glasshouse-grown tomato plants and placed on ice for 4 h in fixing solution under vacuum (300 mm Hg) to remove air bubbles and submerge the entire flower.
- Immunolabeling
- 1 ml of blocking buffer is pipetted onto slide for 2 h (a hydrophobic pen can be used to prevent leakage). Excess solution is dried off the edges of each slide.
- Thereafter, 200 µl of 1:100 LIN5 or INH antibody, diluted in TBST, is pipetted to each slide and incubated for 1 h at RT.
- Slides are then washed 3 x 20 min with TBST. Once again excess solution is blotted from slide with lint free paper (without touching sections) and secondary anti-rabbit IgG fluorescein isothiocyanate (FITC) conjugated antibody, produced in goat is applied at a dilution of 1:200 in TBST incubated for 1 h at RT as above in dark conditions.
- Slides are washed as above with successive wash steps in TBST and then ddH2O (Figure 1A).
- 1 ml of blocking buffer is pipetted onto slide for 2 h (a hydrophobic pen can be used to prevent leakage). Excess solution is dried off the edges of each slide.
- Counterstaining
- A novel counterstaining procedure was developed to ascertain cellular localisation of LIN5 and INH.
- Following immunolabelling, 200 µl of aniline blue is applied to the washed sections and incubated for 20 min in dark conditions.
- Excess aniline blue is washed off by slowly pipetting 5 ml TBST over sections while holding at a 45° and then sections are mounted in 20 μl mowiol-phenylenediamine (mowiol) before a 22 x 50 mm long coverslip is applied.
- Sections are viewed under a ZEISSTM AxioScope. A1 epifluorescence compound microscope. Immunolabelling is viewed using 450-490 nm excitation, long pass 515 nm emission FITC filter set with at 100x objective under oil emersion (Figure 1A).
- Once immunolabelling signal is detected, rapid switching to 365 nm excitation, short pass 420 nm emission UV filter set is achieved to detect fluorescent callose upon binding to aniline blue, as the latter fades rapidly. The image is captured on a ZEISSTM AxioCam digital camera using AxioVision V4.8 software (Figure 1B). Filter set is returned to FITC and another image was taken of immunolabelling in the same target area (Figure 1A).
- When removing each slide from the microscope stage, care must be taken to keep the stage in the same position and to note the orientation of the slide.
- Once slide had been removed, the coverslip was slid off the slide with extreme care and discarded.
- Sections are washed with TBST as above until all excess mowiol is removed and 200 µl of 1:500 4’,6-diamidino-2-phenylindole (DAPI) is applied to locate the nucleolus and incubated at RT for 30 sec.
- Excess DAPI is removed with TBST and slides re-mounted as described above.
- The target area is re-acquired and aligned whilst being viewed under the FITC filter and compared to the previously captured image (Figure 1A) before being switching to long pass 420 nm emission UV filter set and capturing the image of DAPI stained cells (Figure 1C).
- As with DAPI counterstaining procedure, Calcofluor white staining proceeds as above to label cell wall cellulose (Figure 1D).
- All images are then exported and overlayed using Abode Photoshop CS4 software package (Figure 1E).
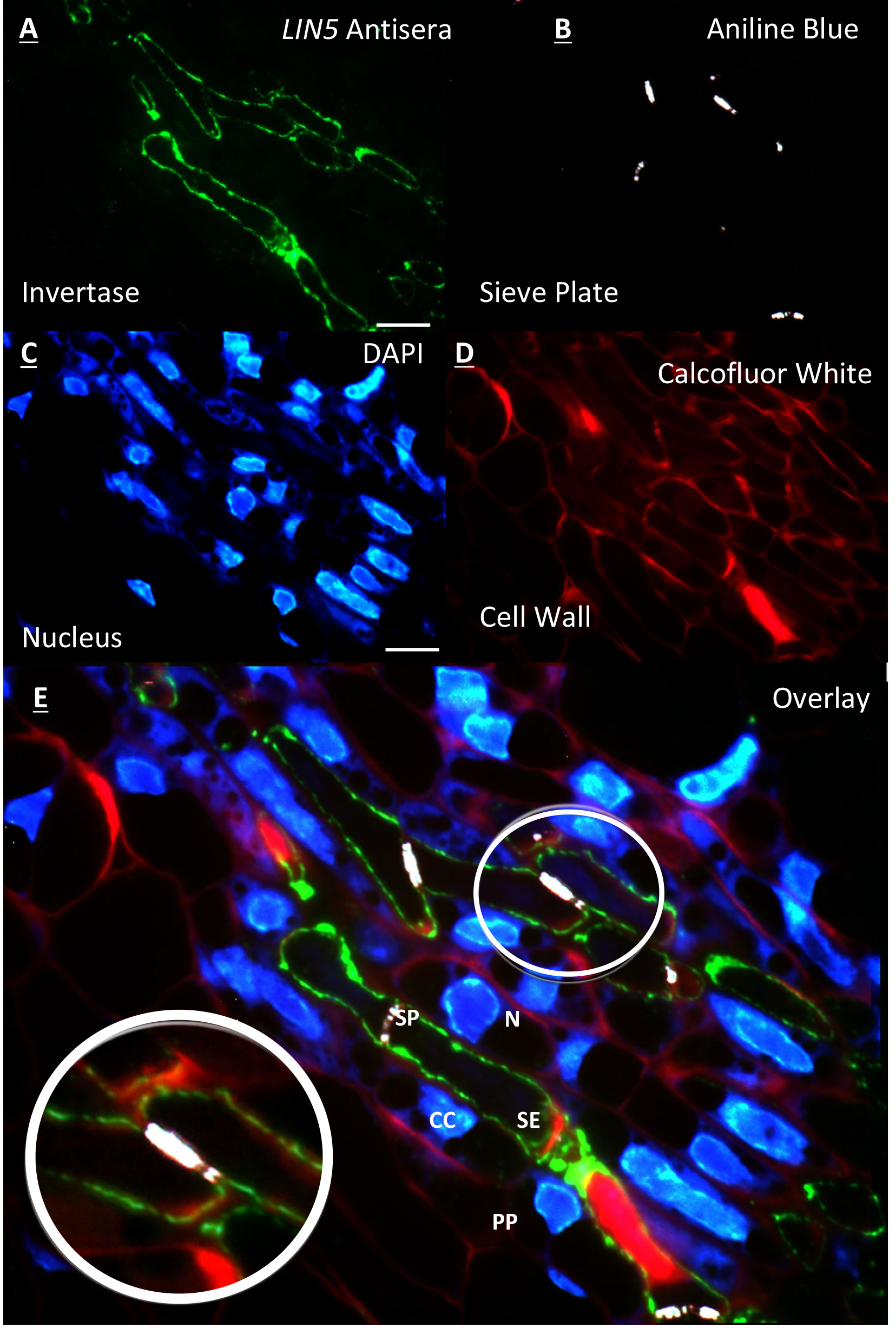
Figure 1. Resin embedded tomato flower section of the placental vascular region 2 DBF. A. Immunolabelling of LIN5 antisera on the cell wall of the sieve element; B. Counterstaining of same section with aniline blue showing the sieve plate at anterior and posterior ends of the sieve element; C. Further counterstaining of same section with DAPI showing nucleus of surrounding cells and no nucleus within sieve elements; D. False coloured red image of cell walls stained with calcofluor white; E. Overlay of A, B, C and D. [zoom] overlay showing LIN5 localisation within the cell wall of the sieve element. All images acquired on a ZEISS compound fluorescence microscope at 100x objective. Scale bars = 10 µm. labelled cell types = (N) nucleus, (CC) companion cell, (PP) phloem parenchyma, (SP) sieve plate and (SE) sieve element. (This image was reproduced from Palmer et al., 2015.)
- A novel counterstaining procedure was developed to ascertain cellular localisation of LIN5 and INH.
Data analysis
Images sequences were imported into Adobe Bridge CS4 software before being loaded into Adobe Photoshop CS4 software and overlaid with opacity and intensity levels adjusted.
Notes
Gelatine coated slides need to be well coated to ensure stability of section on the microscope slide during removal of coverslip for optimal overlaying of images acquired.
Recipes
- Fixing solution
50 mM PIPES, pH 6.8
2% paraformaldehyde
1.5% glutaraldehyde
2 mM CaCl2 - Blocking buffer
20 mM Tris, pH 7
150 mM NaCl
0.2% NaN3
2% bovine serum albumin
Acknowledgments
The condensed version of this method was originally published in Palmer et al., 2015.
This work was collectively supported by the National Science Foundation of China (Grant number 30425043 to Y.L.R.) and Australia Research Council (ARC DP110104931 to Y.L.R. and DP120104148 to Y.L.R. and J.W.P.).
References
- Palmer, W. M., Ru, L., Jin, Y., Patrick, J. W. and Ruan, Y. L. (2015). Tomato ovary-to-fruit transition is characterized by a spatial shift of mRNAs for cell wall invertase and its inhibitor with the encoded proteins localized to sieve elements. Mol Plant 8(2): 315-328.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Palmer, W. M., Patrick, J. W. and Ruan, Y. (2017). Resin-embedded Thin-section Immunohistochemistry Coupled with Triple Cellular Counterstaining. Bio-protocol 7(7): e2052. DOI: 10.21769/BioProtoc.2052.
Category
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Fixed-tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









