- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
PNGase Sensitivity Assay to Study the Folding Status of Proteins
Published: Vol 6, Iss 19, Oct 5, 2016 DOI: 10.21769/BioProtoc.1952 Views: 8038
Reviewed by: Ralph BottcherJoëlle SchlapferGregory C. Finnigan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Novel Protein-oligonucleotide Conjugation Method Involving a High-affinity Capture HaloTag
Junshi Yazaki
Sep 20, 2020 5554 Views

Bioorthogonal Labeling and Chemoselective Functionalization of Lung Extracellular Matrix
Zihan Ling [...] Xi Ren
Feb 20, 2021 4386 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3415 Views
Abstract
This protocol aims to evaluate folding status of proteins, utilizing peptide:N-glycanase (PNGase) sensitivity. In the cytosol, PNGase works as a deglycosylation-enzyme. N-glycans on unfolded/misfolded proteins are more susceptible to PNGase than N-glycans on folded proteins because of the preference of PNGase to non-native proteins. PNGase is endogenously expressed in various cell types, including HCT116 cells, DT40 cells and mouse embryonic fibroblast cells. Partial deglycosylation by PNGase can be detected by faster migration of band in SDS-PAGE. You can compare tightness of the folding among wild-type and mutant proteins of interest. This method can be used with regular molecular and cell biology equipment, but applied only to glycoproteins.
Keywords: PNGaseMaterials and Reagents
- 6 well dish (Corning, Falcon®, catalog number: 353046 ) (Stored at room temperature)
- PVDF membrane (GE Healthcare, catalog number: 10600023 ) (Stored at room temperature)
- DT40 cell line (DT40 is a B cell line derived from an avian leukosis virus induced bursal lymphoma in a white leghorn chicken) (ATCC, catalog number: CRL-2111 ) (Endogenous expression of cytosolic PNGase)
- Homo sapiens colon colorectal carcinoma cell line (HCT116) (ATCC, catalog number: CCL-247 ), HCT116 cells (ATCC, catalog number: CCL-247) (Endogenous expression of cytosolic PNGase)
- Opti-mem (Thermo Fisher Scientific, GibcoTM, catalog number: 31985-070 )
- Lipofectamine 2000 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668019 )
- Dulbecco’s modified Eagle’s medium (DMEM) (NACALAI TESQUE, catalog number: 08458-45 ) (Stored at 4 °C)
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270-106 )
- 100 U/ml penicillin and 100 μg/ml streptomycin (NACALAI TESQUE) (Stored at -20 °C)
- RPMI (NACALAI TESQUE, catalog number: 30263-95 ) (Stored at 4 °C)
- Chicken serum (Thermo Fisher Scientific, GibcoTM, catalog number: 16110-082 )
- 1 M dithiothreitol (DTT) (Wako Pure Chemical Industries, catalog number: 041-08976 ) (for reduction of proteins; in water; stored at -20 °C)
- 20 mM carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (Z-VAD-fmk) (Promega, catalog number: G7231 ) (for inhibition of PNGase activity; stored at -20 °C)
- Nonidet P-40 (NACALAI TESQUE, catalog number: 23640-94 ) (for HCT116; stored at room temperature)
- 4% digitonin (Wako Pure Chemical Industries, catalog number: 043-21371 ) (for DT40; in water; stored at -80 °C)
- Anti-myc antibody (MEDICAL & BIOLOGICAL LABORATORIES, catalog number: 562 ) (Stored at -20 °C)
- Anti-β-actin antibody conjugated with HRP (Wako Pure Chemical Industries, catalog number: 017-24573 )
- NaCl (Wako Pure Chemical Industries, catalog number: 191-01665 )
- Na2HPO4
- KCl
- KH2PO4
- Tris/HCl, pH 8.0 (Sigma-Aldrich, catalog number: T6791 ) (Stored at room temperature)
- Glycerol
- Bromophenol blue (BPB)
- Sodium dodecyl sulfate
- Protease inhibitor cocktail (100x) (NACALAI TESQUE, catalog number: 25955-11 ) (for inhibition of various proteases’ activity; in ddH2O; stored at -20 °C)
- 10 mM Z-Leu-Leu-Leu-CHO (MG132) (PEPTIDE INSTITUTE, catalog number: 3175-v ) (for inhibition of proteasomal activity; in DMSO; stored at -20 °C)
- Phosphate buffered saline (PBS) (see Recipes)
- 2x sodium dodecyl sulfate (SDS) sample buffer (pH 6.8) (see Recipes)
- Buffer A (Stored at 4 °C) (see Recipes)
- Buffer B (see Recipes)
Equipment
- High speed refrigerated micro centrifuge (Tomy, model: MX-301 )
- mPAGE (ATTO, model: AE-6530 ) (Using hand-made 10% gel)
- Transfer equipment (ATTO, model: WSE-4020 )
- Heat block (TAITEC, model: DTU-1BN )
- Micro porator (Digital Bio, model: MP-100 )
Software
- ImageJ
Procedure
- Culture adherent HCT116 cells in Dulbecco’s modified Eagle’s medium (glucose 4.5 g/L) supplemented with 10% fetal bovine serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 37 °C in a humidified 5% CO2/95% air atmosphere. Culture suspended DT40 cells at a density of 1 x 105-1 x 106 cells per ml in RPMI1640 medium supplemented with 10% fetal bovine serum, 1% chicken serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 39.5 °C in a humidified 5% CO2/95% air atmosphere.
- Transfect 8.0 x 105 HCT116 cells (40-60% confluent in 6-well plate) using 1 μg of plasmid DNA, 300 μl of Opti-mem and 10 μl of Lipofectamine 2000 (Invitrogen) for one well according to the manufacturers’ instructions (After the transfection medium is not replaced). To obtain 1.05 x 107 cells transfected cells, electroporate three times 3.5 x 106 DT40 cells with 8 µg plasmid DNA with two pulses at 1,500 V for 15 msec according to the manufacturer’s instructions.
- Collect approximately 2.0 x 106 HCT116 cells or 4.0 x 106 DT40 cells for further analysis 24 h (HCT116 cell) or 16 h (DT40 cells) after transfection. For this the cells are washed with 1 ml PBS three times, scraped off, centrifuged at 800 x g for 2 min at 4 °C. After this step, samples from HCT116 cells and DT40 cells are treated in the same manner.
- Remove supernatants of samples, suspend cell pellets in 200 μl buffer A and incubate for 20 min on ice to lyse the cells.
Note: The buffer A must NOT contain Z-VAD-fmk as it would inihibit PNGase activity. - Clarify cell lysates by centrifugation at 17,800 x g for 10 min at 4 °C and transfer the supernatants to new tubes. Pellets are discarded.
- To allow PNGase processing of misfolded proteins, incubate the cell lysates further for various periods (a good starting range would be 4 h) on ice. Remove 40-50 µl aliquots at different times, immediately mix with 40-50 μl of buffer B supplemented with 100 mM DTT (for reduction) and 2 µM Z-VAD-fmk (to inhibit the PNGase) and incubate at 100 °C for 5 min. Separate 10 µl of the samples by SDS-PAGE followed by immunoblotting using PVDF membrane and probing with an antibody against the protein of interest. Percentage of SDS-PAGE depends on the molecular weight of the protein of interest. Band intensities can be analyzed and compared to each other by ImageJ or similar programs (Figure 1).
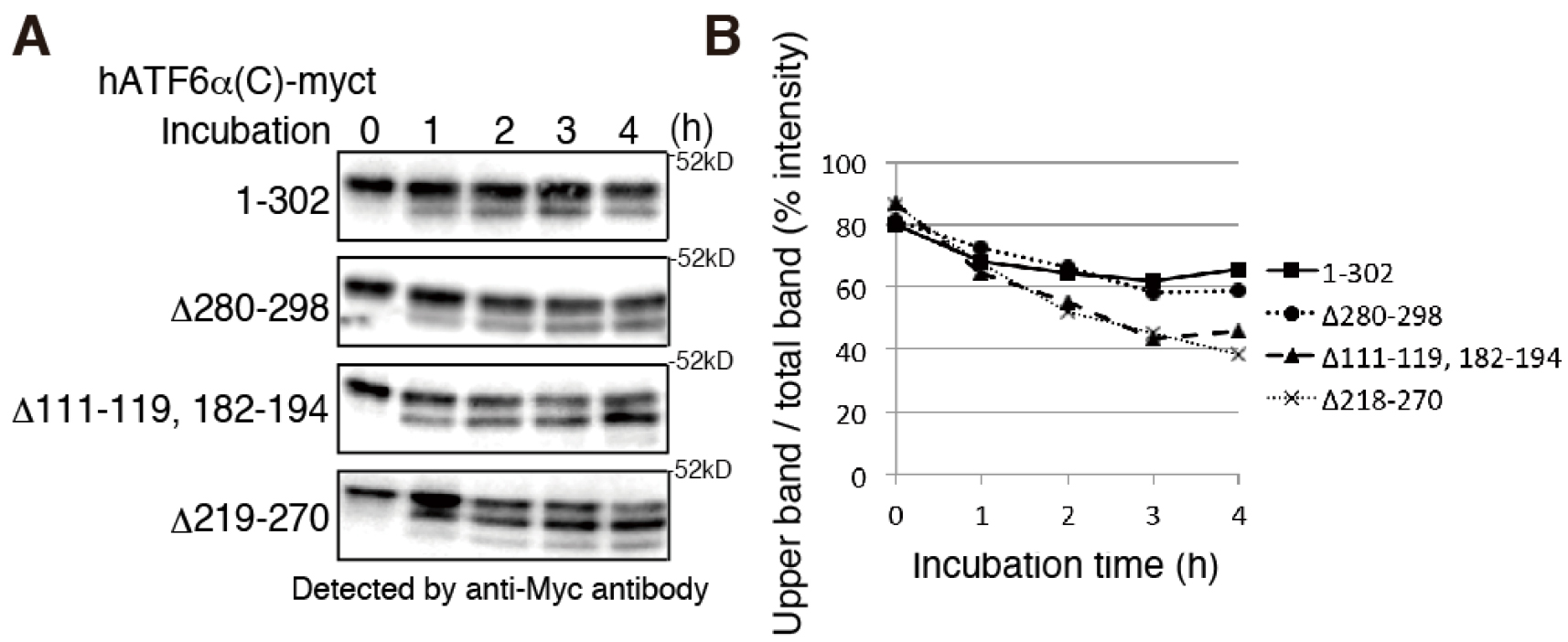
Figure 1. Severely misfolded mutants were more sensitive to endogenous PNGase. A. Chicken EDEM1/2/3 triple KO cells (gEDEM TKO) were described previously (Ninagawa et al., 2015). The indicated myc-tagged hATF6α(C) variants (indicated on the left side of the blots) were transiently expressed in gEDEM-TKO cells and subjected to PNGase sensitivity assay. After the indicated time points the PNGase reaction was stopped and samples analyzed by a 12% SDS-PAGE followed by immunoblotting with antibodies against the myc-tag. The data shown represents a single representative experiment out of two repeats. Less folded proteins are more sensitive to PNGase, so partially deglycosylated bands were detected at earlier time points in hATF6α(C)-myct mutants, Δ111-119, 182-194 mutant and Δ219-270 mutant. See Ninagawa et al. (2015) for further proteins’ information. B. The graph shows % intensity of upper band/total band of (A). The band intensity was analyzed by imageJ.
Notes
- Most cells including HCT116 cells, DT40 cells and mouse embryonic fibroblast cells express PNGase endogenously. Therefore it is not necessary to express PNGase exogenously. Other cell types have to be tested for PNGase expression. PNGase is also expressed in yeast, but we have not checked whether this assay is applicable to yeast cells.
- This assay has been used to determine the folding state of several proteins, however, some glycoproteins are not sensitive to PNGase at all.
- The use of strong detergents, such as SDS, is not appropriate for this method as PNGase activity can be lost. In our case, we used 1% NP-40 for mammalian cells and 1% digitonin for chicken cells. But you can use 1% NP-40 for chicken cells and 1% digitonin for mammalian cells. Not too strong detergent can be used for PNGase assay.
- As control one can include two micromole of the PNGase inhibitor Z-vad-fmk.
- You can normalize the sample by the concentration of proteins or cell number. Or you can use anti-β-actin antibody conjugated with HRP for SDS-PAGE to show the equal load of proteins.
- We can provide plasmids to express hATF6α(C)-myct 1-302 and Δ280-298 for positive controls (Folded), and Δ111-119, 182-194 and Δ219-270 for negative controls (Less folded).
- This assay has been used for exogenously expressed proteins, but should also be applicable to endogenous proteins.
- We recommend that when you analyze glycoproteins, you should include Z-VAD-fmk in lysis buffer in order to inhibit PNGase activity after cell lysis.
- Recombinant PNGase is also available for PNGase sensitivity assay (Liu et al., 2016).
Recipes
- Phosphate buffered saline (PBS)
137 mM NaCl
8.1 mM Na2HPO4
2.68 mM KCl
1.47 mM KH2PO4 - 2x sodium dodecyl sulfate (SDS) sample buffer (pH 6.8)
100 mM Tris/HCl (pH 6.8)
20% glycerol
0.2% bromophenol blue (BPB)
4% sodium dodecyl sulfate - Buffer A
50 mM Tris/HCl (pH 8.0)
1% NP-40 or 1% digitonin
150 mM NaCl
Protease inhibitor cocktail (Add before use)
20 μM MG132 (Add before use)
Stored at 4 °C
Note: 1% NP-40 is for mammalian cells such as HCT116 cells and 1% digitonin is for chicken cells such as DT40 cells. - Buffer B
1x SDS sample buffer (Stored at room temperature)
100 mM dithiothreitol (Add before use)
2 μM Z-VAD-fmk (Add before use)
Acknowledgments
This protocol was adapted from and used in Ninagawa et al. (2015).
References
- Liu, Y. C, Fujimori, D. G. and Weissman, J. S. (2016). Htm1p-Pdi1p is a folding-sensitive mannosidase that marks N-glycoproteins for ER-associated protein degradation. Proc Natl Acad Sci U S A. 113(28): E4015-4024.
- Ninagawa, S., Okada, T., Sumitomo, Y., Horimoto, S., Sugimoto, T., Ishikawa, T., Takeda, S., Yamamoto, T., Suzuki, T., Kamiya, Y., Kato, K. and Mori, K. (2015). Forcible destruction of severely misfolded mammalian glycoproteins by the non-glycoprotein ERAD pathway. J Cell Biol 211(4): 775-784.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ninagawa, S. and Mori, K. (2016). PNGase Sensitivity Assay to Study the Folding Status of Proteins. Bio-protocol 6(19): e1952. DOI: 10.21769/BioProtoc.1952.
- Ninagawa, S., Okada, T., Sumitomo, Y., Horimoto, S., Sugimoto, T., Ishikawa, T., Takeda, S., Yamamoto, T., Suzuki, T., Kamiya, Y., Kato, K. and Mori, K. (2015). Forcible destruction of severely misfolded mammalian glycoproteins by the non-glycoprotein ERAD pathway. J Cell Biol 211(4): 775-784.
Category
Biochemistry > Protein > Modification
Biochemistry > Carbohydrate > Glycoprotein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










