- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Non-radioactive in vitro PINK1 Kinase Assays Using Ubiquitin or Parkin as Substrate
Published: Vol 6, Iss 19, Oct 5, 2016 DOI: 10.21769/BioProtoc.1946 Views: 9117
Reviewed by: Ralph BottcherQiangjun ZhouVanesa Olivares-Illana

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins
Nitu Saha [...] Mark Hochstrasser
Sep 5, 2025 3785 Views

Verification of N-Linked Glycosylation of Proteins Isolated from Plant or Mammalian Cell Lines Using PNGase Enzyme
Wuqiang Hong [...] Jiayang Li
Sep 20, 2025 1918 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 483 Views
Abstract
This protocol describes the in vitro phosphorylation of ubiquitin and Parkin by the kinase PINK1 using recombinant proteins. Both substrates, ubiquitin and Parkin, are phosphorylated at the conserved serine 65 residue (pS65-ubiquitin and pS65-Parkin). The protocol also includes the use of monomeric and K48- and K63-linked poly-ubiquitin chains as alternative substrates. Although there are commercially available antibodies, we have not tested their performance in this assay since, but used validated antibodies from our laboratory. An alternative antibody-independent method, the use of phos-tag gels to detect pS65-ubiquitin and pS65-Parkin, is described in addition.
Keywords: PINK1Background
In cells, PINK1 is stabilized and activated by mitochondrial membrane depolarization and other forms of stress that lead to mitochondrial damage. Activated PINK1 phosphorylates ubiquitin, which acts as the receptor for the cytosolic E3 ubiquitin ligase Parkin on the mitochondrial surface. Phosphorylation of Parkin by PINK1 is required for full activity of Parkin towards mitochondrial substrates. The presence of active pS65-Parkin amplifies in a feed-forward mechanism the amount of pS65-ubiquitin on mitochondria, which acts as the mitophagy tag. Eventually, damaged mitochondria are being recognized by autophagy adapters and will be degraded by the proteasome and by autophagy (mitophagy). This crucial mitochondrial quality control pathway promotes turnover of mitochondria and prevents accumulation of dysfunctional mitochondria that can lead to cellular degeneration. Loss-of-function mutations in either PINK1 or Parkin are associated with early-onset Parkinson’s disease.
In cell-free ubiquitination assays, the presence of pS65-Ub activates the E3 ligase activity of Parkin also in the absence of active PINK1. While many groups use their own purified recombinant proteins for in vitro PINK1 kinase assays, this optimized protocol describes the in vitro phosphorylation of ubiquitin and Parkin with commercially available recombinant proteins.
Materials and Reagents
- Recombinant proteins
- Active PINK1 [MBP-tagged] from Tribolium castaneum (Ubiquigent, catalog number: 66-0043-050 )
- Inactive PINK1 D359A [MBP-tagged] from Tribolium castaneum (Ubiquigent, catalog number: 66-0044-050 )
- Parkin (untagged) (Ubiquigent, catalog number: 63-0048-025 )
- Ubiquitin (untagged) (BOSTON BIOCHEM, catalog number: U-100H )
- Ubiquitin N-terminal biotin (BOSTON BIOCHEM, catalog number: UB-560 )
- Biotinylated poly-ubiquitin chains (K48-linked) (BOSTON BIOCHEM, catalog number: UCB-230 )
- Poly-ubiquitin chains (K63-linked) (Biotinylated) (BOSTON BIOCHEM, catalog number: UCB-330 )
- Active PINK1 [MBP-tagged] from Tribolium castaneum (Ubiquigent, catalog number: 66-0043-050 )
- Chemicals for buffers
- HEPES (Sigma-Aldrich, catalog number: H4034 )
- DL-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- EDTA (EMD Millipore, catalog number: EX0539 )
- ATP (AppliChem, catalog number: A1348 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M0250 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 60370 )
- Tris (Santa Cruz Biotechnology, catalog number: sc-3715 )
- Hydrochloric acid (37%) (EMD Millipore, catalog number: 1003172500 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L4509 )
- Glycerol (Thermo Fisher Scientific, Fisher Scientific, catalog number: BP2291 )
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- Bromophenol blue sodium salt (Sigma-Aldrich, catalog number: B5525 )
- Glycine (Sigma-Aldrich, catalog number: G7126 )
- Methanol (Pharmco-Aaper, catalog number: 339000000 )
- Dry milk powder
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A9647 )
- Sodium chloride (NaCl) (Thermo Fisher Scientific, Fisher Scientific, catalog number: BP358-10 )
- Tween-20 (Sigma-Aldrich, catalog number: P1379 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P3911 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S5136 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655 )
- 10x kinase buffer (see Recipes)
- 6x SDS loading buffer (see Recipes)
- SDS running buffer (see Recipes)
- Blotting buffer (see Recipes)
- TBST (see Recipes)
- Resolving gel buffer (see Recipes)
- 14% resolving phos-tag gel (recipe for mini gel) (see Recipes)
- 8% resolving phos-tag gel (recipe for mini gel) (see Recipes)
- Stacking gel buffer (see Recipes)
- Stacking gel (recipe for two mini gels) (see Recipes)
- Phos-tag gel washing buffer 1 (see Recipes)
- Phos-tag gel washing buffer 2 (see Recipes)
- Phosphate buffer saline (PBS) (see Recipes)
- Antigen retrieval solution (see Recipes)
- HEPES (Sigma-Aldrich, catalog number: H4034 )
- SDS-PAGE/Western blot
- 8-16% Tris-glycine gels (for Parkin) (Thermo Fisher Scientific, InvitrogenTM, catalog number: EC60485BOX )
- 16% Tris-glycine gels (for ubiquitin) (Thermo Fisher Scientific, InvitrogenTM, catalog number: EC64985BOX )
- Pre-stained protein ladder (Bio-Rad Laboratories, catalog number: 1610394 )
- PVDF membrane (EMD Millipore, catalog number: IPVH00010 )
- pS65-Parkin antibody (Abcam, catalog number: ab154995 )
- pS65-Ubiquitin antibody (EMD Millipore, catalog number: ABS1513-I or BOSTON BIOCHEM, catalog number: A-110 )
- Monoclonal Parkin antibody Prk8 (Cell Signaling Technology, catalog number: 4211 )
- Monoclonal Ubiquitin antibody (LifeSensor, catalog number: VU-101 )
- PierceTM streptavidin, HRP-linked (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 21130 )
- HRP-conjugated anti-rabbit antibodies (Jackson Immunoresearch, catalog number: 111-035-003 )
- HRP-conjugated anti-mouse antibodies (Jackson Immunoresearch, catalog number: 115-035-003 )
- Western blot detection reagent (EMD Millipore, catalog number: WBKLS0500 )
- X-ray films (Genesee Scientific, catalog number: 30-101L )
- Acrylamide:Bis-acrylamide 19:1, solution 40% (w/v) (EMD Millipore, catalog number: 1290-OP )
- Zinc chloride (ZnCl2) (Sigma-Aldrich, catalog number: 96468 )
- Ammonium persulfate [(NH4)2S2O8] (Sigma-Aldrich, catalog number: A9164 )
- TEMED (Santa Cruz Biotechnology, catalog number: sc-29111 )
- Phos-tagTM acrylamide AAL-107 (Wako Pure Chemical Industries, catalog number: 304-93521 )
- 25% glutaraldehyde solution (Sigma-Aldrich, catalog number: G6257 )
- 8-16% Tris-glycine gels (for Parkin) (Thermo Fisher Scientific, InvitrogenTM, catalog number: EC60485BOX )
Equipment
- Standard bench-top centrifuge
- Heated shaker for microtubes [e.g., Thermomixer (Eppendorf, catalog number: 5384000020 )]
- SDS-PAGE gel electrophoresis and blotting boxes (e.g., VWR International, Peqlab, catalog number: 45-1010-C and Bio-Rad Laboratories, catalog number: 1704070 )
- Orbital shaker for Western blots
- X-ray film processor or camera detection system (e.g., GE Healthcare, catalog number: 28-9558-10 ) to visualize chemiluminescent Western blot signal
Software
- ImageJ or ImageStudio Lite
Procedure
- Kinase reaction
- Prepare 10x kinase buffer.
Note: 10x kinase buffer should always be prepared freshly. It can be mixed together from higher concentrated stock solutions. We recommend freezing small (one time use) aliquots of ATP and DTT. - Thaw recombinant proteins on ice.
- In a clean microtube, mix PINK1 with ubiquitin (monomers or chains) or Parkin: use 0.1-1 µg of PINK1 per µg of ubiquitin or Parkin.
Notes:- We recommend using 1 µg of substrate in a 20 µl reaction as a starting point. We typically use 20-100 µl as a total reaction volume.
- If biotinylated ubiquitin is used, HRP-coupled streptavidin can be used for detection on phos-tag gels. Otherwise an ubiquitin antibody is necessary.
- Compared to poly-ubiquitin chains, free monomeric ubiquitin is simpler to analyze on phos-tag gels since there is only one band (and therefore only one band-shift). While polymeric K48- and K63-linked chains represent physiological substrates of PINK1 (when attached to mitochondrial substrate proteins), phosphorylated and non-phosphorylated bands may at least partially overlap in particular if chains of various lengths are used.
- We recommend using 1 µg of substrate in a 20 µl reaction as a starting point. We typically use 20-100 µl as a total reaction volume.
- Optional: Prepare negative control sample as above but use inactive PINK D359A instead of the wild-type enzyme.
- Add kinase buffer to a final concentration of 1x.
- Spin reactions for 10 sec at 1,000 x g at 4 °C.
- Incubate the reactions for 60-120 min (or 24-48 h to achieve 100% substrate phosphorylation) in a heated shaker at 37 °C.
- Stop the reactions by adding 6x SDS loading buffer to a final concentration of 1x.
- Incubate the reactions for 15 min at 56 °C.
Note: This lower temperature prevents the unwanted spontaneous formation of ubiquitin 'dimers'.
- Prepare 10x kinase buffer.
- Detection using phospho-specific antibodies
- Assemble SDS gel into an electrophoresis chamber.
- Fill the chamber with 1x SDS running buffer.
- Load samples on the gel. Use 20-500 ng per lane.
Note: As a starting point we recommend using 50 ng. Depending on the detection method (antibody, dilution etc.) and equipment the amounts may be changed. - Add 5-10 µl of pre-stained protein ladder in one lane as molecular weight marker.
- Run gel using standard conditions (for the gels/equipment listed below use constant 125 Volt for 90 min).
- Transfer gel onto methanol-activated PVDF membrane in 1x blotting buffer using standard conditions (for the gels/equipment listed below use constant 100 Volt for 50 min).
- Block membrane in 5% dry milk powder in TBST for 1 h at room temperature, shaking.
Note: We routinely use TBST for Western blots. Other wash buffers might work as well, however phosphate-based buffers are generally not recommended for the detection of phosphorylated proteins. - Incubate membrane with phospho-specific ubiquitin or Parkin antibodies diluted in 5% BSA in TBST overnight at 4 °C.
Note: The phospho-specific antibodies listed below have not been tested for this particular method. The final concentration of antibody will depend on the amount loaded and must be determined empirically. - Wash membrane 4 x 10 min in TBST at RT.
- Incubate membrane with secondary antibodies diluted 1:10,000 in 5% dry milk powder in TBST for one hour at RT.
- Wash membrane 4 x 10 min in TBST at RT.
- Incubate membrane with detection reagent according to manufacturer’s instructions.
- Develop Western blot by exposing X-ray films or using a camera detection system.
- Assemble SDS gel into an electrophoresis chamber.
- Detection using phos-tag gels
Note: This method is independent of the availability/performance of phospho-specific antibodies. Phosphorylated proteins migrate slower in phos-tag gels compared to non-modified proteins, resulting in a band shift. They can be visualized with specific antibodies against the unmodified protein of interest or with HRP-coupled streptavidin if biotinylated recombinant substrates were used for the kinase reaction.- Prepare phos-tag polyacrylamide gels of desired density: 8% for Parkin and 14% for mono ubiquitin, respectively.
- Proceed with SDS-PAGE as above.
Notes:- Run gels in ice or with cold water perfusion of the apparatus [for the equipment listed below use constant 125 Volt for 90 min (8% gel) and 120-140 min (14% gel), respectively].
- Pre-stained protein markers generally cause distortion because they contain EDTA or phosphorylated proteins. At least one lane should be left blank between the marker and the samples. Please note that the molecular weight of pre-stained markers will not be accurate on phos-tag gels.
- Run gels in ice or with cold water perfusion of the apparatus [for the equipment listed below use constant 125 Volt for 90 min (8% gel) and 120-140 min (14% gel), respectively].
- After electrophoresis incubate phos-tag gels in phos-tag washing buffer 1 for 20 min followed by two incubations, each 20 min, in phos-tag washing buffer 2 at RT.
- Transfer gel onto methanol-activated PVDF membrane in 1x blotting buffer (for equipment listed below use constant 105 Volt for 60 min).
Note: Nitrocellulose membrane might be used instead of PVDF. - For some primary antibodies, antigen retrieval is necessary. Ubiquitin antibody VU-1 requires washing of the membrane with ultrapure water prior to 0.5% glutaraldehyde treatment for 20 min, shaking, followed by three brief washes with PBS.
- Block membrane for 1 h at room temperature in 5% dry milk powder in TBST for VU-1 and Prk8 antibodies and in 5% BSA in TBST for streptavidin linked to HRP. Proceed with Western blot as above.
Note: For 50 ng of substrate use a 1:5,000 dilution of VU-1 antibody to detect ubiquitin and 1:150,000 dilution of Prk8 antibody to detect Parkin. In case of biotinylated ubiquitin, use streptavidin-HRP at a dilution of 1:100,000. For other amounts, dilution of antibodies should be adjusted accordingly. For dilutions > 1:50,000 a predilution is recommended (e.g., dilute antibody 1:100 and then 1:1,000 for a 1:100,000 dilution). If sodium azide is added predilutions can be stored at 4 °C. However, sodium azide will inhibit HRP and is therefore not recommended for streptavidin-HRP.
- Prepare phos-tag polyacrylamide gels of desired density: 8% for Parkin and 14% for mono ubiquitin, respectively.
Data analysis
The here described method uses Western blot analysis as read-out. Phosphorylation by PINK1 over time (Figure 1) can be quantified using densitometric analysis of band intensities using open source software (e.g., ImageJ or ImageStudio Lite). Reactions performed with mutant PINK1 D359A serve as a negative control. On phos-tag gels (Figure 1) the ratio of the shifted versus the non-shifted band allows to calculate the ratio of modified to non-modified substrate. Statistical analysis should be performed with the results from at least three independent experiments.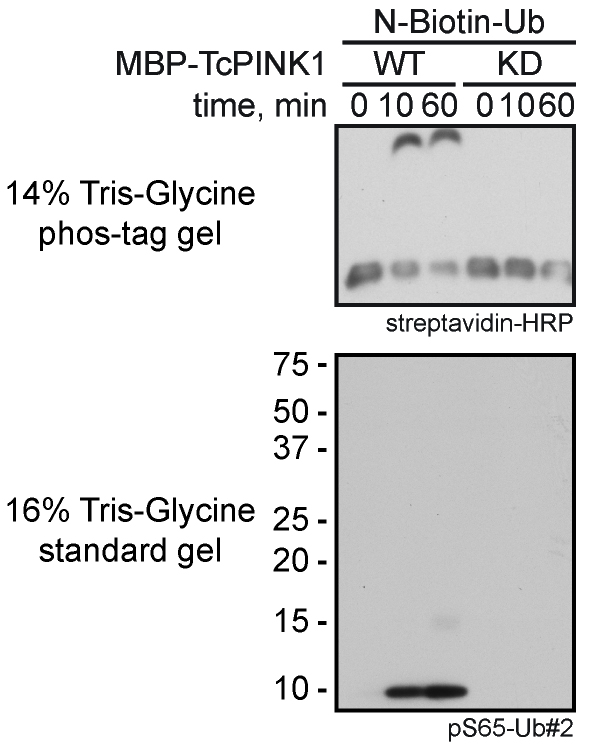
Figure 1. Kinase assay with N-terminally tagged monomeric ubiquitin using wild-type (WT) or kinase-dead (KD) MBP-TcPINK1. Phosphorylated ubiquitin was detected on a 14% Tris-glycine phos-tag gel with HRP-coupled streptavidin (upper panel) and on a standard 16% Tris-glycine gel with a phospho-specific ubiquitin antibody (pS65-Ub#2) (lower panel).
Recipes
- 10x kinase buffer
Note: Prepare freshly.
200 mM HEPES, pH 7.4
100 mM DTT
1 mM EGTA
1 mM ATP
100 mM MgCl2 - 6x SDS loading buffer
Note: Store at -20 °C.
375 mM Tris, pH 6.8
9% (w/v) SDS
50% (v/v) glycerol
9% (v/v) β-mercaptoethanol
0.03% (w/v) bromophenol blue - SDS running buffer
Note: Store at RT.
25 mM Tris
200 mM glycine
0.1% (w/v) SDS - Blotting buffer
Note: Store at 4 °C.
25 mM Tris
192 mM glycine
0.1% (w/v) SDS
20% (v/v) methanol - TBST
Note: Store at RT.
50 mM Tris, pH 7.4
150 mM NaCl
0.1% (v/v) Tween-20 - Resolving gel buffer
Note: Store at 4 °C.
1.5 M Tris-Cl, pH 8.8
0.4% (v/v) SDS - 14% resolving phos-tag gel (recipe for one mini gel)
Note: Prepare freshly.
2.75 ml resolving gel buffer
3.875 ml AA:Bis 19:1, solution 40% (w/v)
4.103 ml water
111 µl 5 mM phos-tag (final concentration: 50 µM)
111 µl 10 mM ZnCl2 (final concentration: 100 µM)
110 µl 10% (w/v) ammonium persulfate in water
11 µl TEMED - 8% resolving phos-tag gel (recipe for one mini gel)
Note: Prepare freshly.
2.75 ml resolving gel buffer
2.214 ml AA:Bis 19:1, solution 40% (w/v)
5.764 ml MQ water
111 µl 5 mM phos-tag (final concentration: 50 µM)
111 µl 10 mM ZnCl2 (final concentration: 100 µM)
110 µl 10% (w/v) ammonium persulfate in MQ water
11 µl TEMED - Stacking gel buffer
Note: Store at 4 °C.
0.5 M Tris-Cl, pH 6.8
0.4% (w/v) SDS - Stacking gel (recipe for two mini gels)
Note: Prepare freshly.
2.06 ml stacking gel buffer
930 µl AA:Bis 19:1, solution 40% (w/v)
5.17 ml MQ water
83 µl 10% (w/v) ammonium persulfate in MQ water
8 µl TEMED - Phos-tag gel washing buffer 1
Note: Prepare freshly or store at RT.
1x blotting buffer
0.01% (w/v) SDS
1 mM EDTA, pH 8.0 - Phos-tag gel washing buffer 2
Note: Prepare freshly or store at RT.
1x blotting buffer
0.01% (w/v) SDS - Antigen retrieval solution
Note: Prepare freshly from 25% (v/v) glutaraldehyde solution or store at RT.
PBS
0.5% (v/v) glutaraldehyde - Phosphate buffer saline (PBS)
Note: Store at RT.
137.93 mM NaCl
2.67 mM KCl
8.06 mM Na2HPO4
1.47 mM KH2PO4
Acknowledgments
This protocol was adapted from Fiesel et al. (2015) and Fiesel and Springer (2015). FCF is the recipient of a fellowship from the American Parkinson’s Disease Association. WS was partly supported by the NIH/NINDS R01NS085070, the Michael J. Fox Foundation for Parkinson’s Research, the Foundation for Mitochondrial Medicine, the Mayo Clinic Foundation, the Center for Individualized Medicine, the Center for Regenerative Medicine, the Center for Biomedical Discovery, the Marriott Family Foundation, and a Gerstner Family Career Development Award. WS is a member of a Neuroscience-Focused Research Team at Mayo Clinic Florida.
References
- Fiesel, F. C. and Springer, W. (2015). Disease relevance of phosphorylated ubiquitin (p-S65-Ub). Autophagy 11(11): 2125-2126.
- Fiesel, F. C., Ando, M., Hudec, R., Hill, A. R., Castanedes-Casey, M., Caulfield, T. R., Moussaud-Lamodiere, E. L., Stankowski, J. N., Bauer, P. O., Lorenzo-Betancor, O., Ferrer, I., Arbelo, J. M., Siuda, J., Chen, L., Dawson, V. L., Dawson, T. M., Wszolek, Z. K., Ross, O. A., Dickson, D. W. and Springer, W. (2015). (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep 16(9): 1114-1130.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Fiesel, F. C., Hudec, R. and Springer, W. (2016). Non-radioactive in vitro PINK1 Kinase Assays Using Ubiquitin or Parkin as Substrate. Bio-protocol 6(19): e1946. DOI: 10.21769/BioProtoc.1946.
- Fiesel, F. C., Ando, M., Hudec, R., Hill, A. R., Castanedes-Casey, M., Caulfield, T. R., Moussaud-Lamodiere, E. L., Stankowski, J. N., Bauer, P. O., Lorenzo-Betancor, O., Ferrer, I., Arbelo, J. M., Siuda, J., Chen, L., Dawson, V. L., Dawson, T. M., Wszolek, Z. K., Ross, O. A., Dickson, D. W. and Springer, W. (2015). (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep 16(9): 1114-1130.
Category
Neuroscience > Cellular mechanisms > Mitochondria
Biochemistry > Protein > Modification
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link